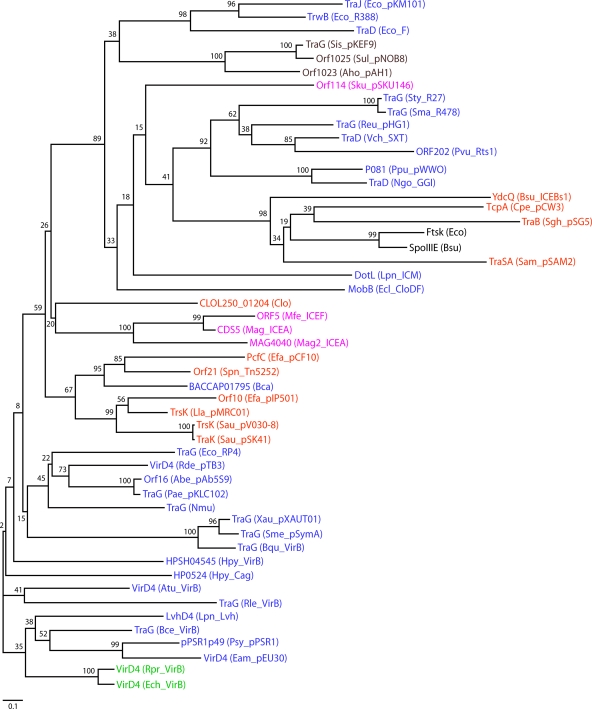
FIG. 2.
Phylogenetic tree of T4CP family members. Sequence alignment and phylogeny estimation were performed using the program MAFFT, version 6.0 (available at http://align.bmr.kyushu-u.ac.jp/mafft/software). The sequence alignment was performed by using the E-INS-i method and default parameters of the program. The tree was constructed using the neighbor-joining method and the following parameters: all ungapped sites from the alignment, JTT amino acid substitution model, and ignore heterogeneity among sites. Bootstrap values for 500 replicates are indicated. The different groups of bacteria are indicated by the following color scheme: blue for gram-negative bacteria, green for gram-negative obligate intracellular pathogens, orange for gram-positive bacteria, pink for cell wall-less bacteria, dark red for Archaea, and black for the related DNA translocases FtsK and SpoIIIE. T4CP designations include the following protein names followed by the species name and plasmid, ICE, or T4SS in parentheses, according to the GenBank database. Accession numbers of T4CPs are AAB58711.1 for Eco_pKM101_TraJ, CAA44852.1 for Eco_R388_TrwB, BAA97972 for Eco_F_TraD, YP_138373.1 for Sis_pKEF9_TraG, CAA09120.1 for Sul_pNOB8_Orf1025, ACI15704.1 for Aho_pAH1_Orf1023, AAS59568.1 for Sku_pSKU146_Orf14, NP_058332.1 for Sty_R27_TraG, NP_941281.1 for Sma_R478_TraG, NP_943001.1 for Reu_pHG1_TraG, AAL59680.1 for Vch_SXT_TraD, NP_640162.1 for Pvu_Rts1_ORF202, NP_542873.1 for Ppu_pWWO_p081, AAW83057.1 for Ngo_GGI_TraD, CAB12293.1 for Bsu_ICEBs1_YdcQ, ABF47325.1 for Cpe_pCW3_TcpA, CAA56759.1 for Sgh_pSG5_TraB, CAA90178.1 for Eco_FtsK, NP_389562.1 for Bsu_SpoIIIE, CAA06449.1 for Sam_pSAM2_TraSA, O54524 for Lpn_ICM_DotL, CAB62409.1 for Ecl_CloDF_MobB, ZP_02074434.1 for Clo_CLOL250_01204, AAN85238.1 for Mfe_ICEF_Orf5, CAJ32610.1 for Mag_ICEA_CDS5, CAL59102.1 for Mag2_ICEA_MAG4040, YP_195789.1 for Efa_pCF10_PcfC, AAG38037.1 for Spn_Tn5252_Orf21, ZP_02036195.1 for Bca_BACCAP01795, CAD44390.1 for Efa_pIP501_Orf10, NP_047302.1 for Lla_pMRC01_TrsK, YP_001653098.1 for Sau_pV030-8_TrsK, NP_863634.1 for Sau_pSK41_TraK, CAA38334.1 for Eco_RP4_TraG, YP_771875.1 for Rde_pTB3_VirD4, YP_001220615.1 for Abe_pAb5S9_Orf16, AAP22624.1 for Pae_pKLC102_TraG, YP_413489.1 for Nmu_TraG, YP_001409435.1 for Xau_pXAUT01_TraG, NP_435748.1 for Sme_pSymA_TraG, YP_032630.1 for Bqu_VirB_TraG, YP_001910374.1 for Hpy_VirB_HPSH04545, NP_207320.1 for Hpy_Cag_HP0524, AAF77174.1 for Atu_VirB_VirD4, ZP_02859218.1 for Rle_VirB_TraG, CAB60062.1 for Lpn_Lvh_LvhD4, ABF79722.1 for Bce_VirB_TraG, NP_940734.1 for Psy_pPSR1_pPSR1p49, NP_943287.1 for Eam_pEU30_VirD4, H71684 for Rpr_VirB_VirD4, and Q8RPL9 for Ech_VirB_VirD4. Species name abbreviations are as follows: Eco, E. coli; Sis, Sulfolobus islandicus; Sul, Sulfolobus sp. strain NOB8H2; Aho, “Acidianus hospitalis”; Sku, S. kunkelii; Sty, S. enterica serovar Typhi; Sma, Serratia marcescens; Reu, Ralstonia eutropha; Vch, V. cholerae; Pvu, Proteus vulgaris; Ppu, Pseudomonas putida; Ngo, N. gonorrhoeae; Bsu, B. subtilis; Cpe, C. perfringens; Sgh, Streptomyces ghanaensis; Sam, Streptomyces ambofaciens; Lpn, L. pneumophila; Eclo, Enterobacter cloacae; Clo, Clostridium sp. strain L2-50; Mfe, M. fermentans; Mag, M. agalactiae strain 5632; Mag2, M. agalactiae PG2; Efa, E. faecalis; Spn, S. pneumoniae; Bca, B. capillosus; Lla, L. lactis; Sau, S. aureus; Rde, Roseobacter denitrificans; Abe, Aeromonas bestiarum; Pae, Pseudomonas aeruginosa; Nmu, Nitrosospira multiformis; Xau, Xanthobacter autotrophicus; Sme, Sinorhizobium meliloti; Bqu, Bartonella quintana; Hpy, H. pylori; Atu, A. tumefaciens; Rle, Rhizobium leguminosarum; Bce, B. cenocepacia; Psy, P. syringae; Eam, Erwinia amylovora; Rpr, R. prowazekii; Ech, E. chaffeensis.