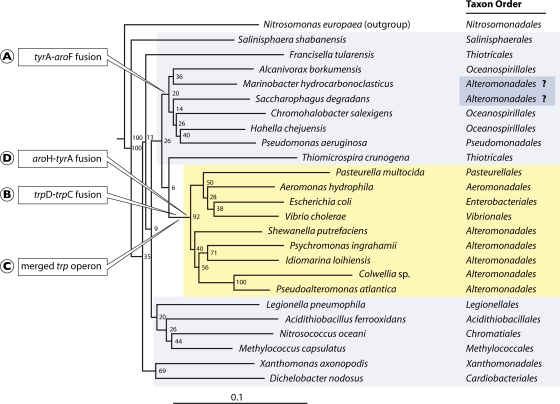
FIG. 3.
16S rRNA tree of Gammaproteobacteria. The TreeBuilder tool at the ribosomal database (http://rdp.cme.msu.edu) was used. The sequence of N. europaea (Betaproteobacteria) was used as an outgroup. The lower-Gammaproteobacteria orders are highlighted in yellow, and the upper-Gammaproteobacteria orders are shown in gray. The evolutionary times for acquisition of four character states (circled letters A, B, C, and D) are shown at appropriate positions on the tree. Fusion A is for genes encoding cyclohexadienyl dehydrogenase (tyrA) and enolpyruvyl-shikimate-3-phosphate synthase (aroF), fusion B is for genes encoding indoleglycerol phosphate synthase (trpD) and phosphoribosyl-anthranilate isomerase (trpC), fusion D is for genes encoding chorismate mutase (aroH) and cyclohexadienyl dehydrogenase (tyrA), and evolutionary event C is the merging of the split-operon components of the trp operon (90). The current designations of two organisms at the upper right as Alteromonadales is considered highly doubtful, as indicated by question marks, because unlike the five Alteromonadales organisms shown in yellow, they possess character state A and they lack character states B, C, and D.