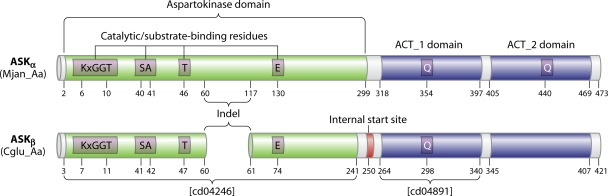
FIG. 6.
Schematic depicting the essential differences between an ASKα enzyme and an ASKβ enzyme. The enzyme selected to represent each subhomology division has been studied by X-ray crystallography and is from M. jannaschii (Mjan_Aa) or C. glutamicum (Cglu_Aa), respectively. Catalytic and substrate-binding residues are shown only as far as the first catalytic residue past the indel region. An unknown query can readily be recognized unambiguously as belonging to ASKα or to ASKβ by obtaining the cd identifier (at the top of the cd hierarchy). If the cd for the Ask domain is cd04246, then the enzyme is in the ASKβ division, because all ASKβ enzymes have that cd (Fig. 7). If the cd identifier is anything else, the enzyme is in the ASKα division, which has five cd numbers (Fig. 7). Likewise, if the cd for the ACT_1 domain is cd04891, it is in the ASKβ division, because all members of that division have this cd. If the cd is anything else, the enzyme belongs to the ASKα division. Internal start sites are shown in Fig. 12 for all ASKβ cohesion group representatives or orphans. The Q…Q motif is shown with one glutamine residue in ACT_1 and the other in ACT_2 for ASKα enzymes that exhibit threonine allostery. A glutamine residue is present in the homologous position of the ACT_1 domain in ASKβ enzymes that are inhibited by threonine or by Thr plus Lys. However, ACT_2 domains of ASKβ enzymes never possess a Q residue in the homologous position.