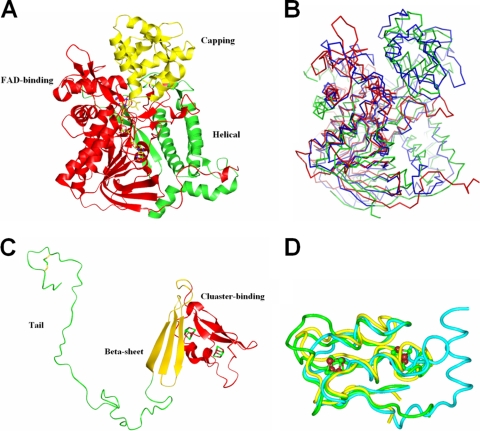
FIG. 4.
Subunit structures of APSR from D. gigas. (A) α-Subunit. The structure of the α-subunit is grouped into the following three parts: the FAD-binding domain (red), capping domain (yellow), and helical domain (green). (B) Superimposition of FAD-binding domains. The structure of the FAD-binding domain of the α-subunit (red) is superimposed with aspartate oxidase (green) and fumarate reductase (blue) from E. coli. The superimposition shows that less similarity is exhibited in the partial FAD-binding domain that is near FAD. (C) β-Subunit. The β-subunit structure is grouped into the following three segments: [4Fe-4S] cluster-binding domain (red), β-sheet domain (yellow), and C-terminal tail domain (green). (D) Structural alignment of the cluster-binding domain in the β-subunit. The cluster-binding domain in the β-subunit of APSR from D. gigas (green) is superimposed with ferredoxin II from D. gigas (yellow) and ferredoxin from Chromatium vinosum (blue).