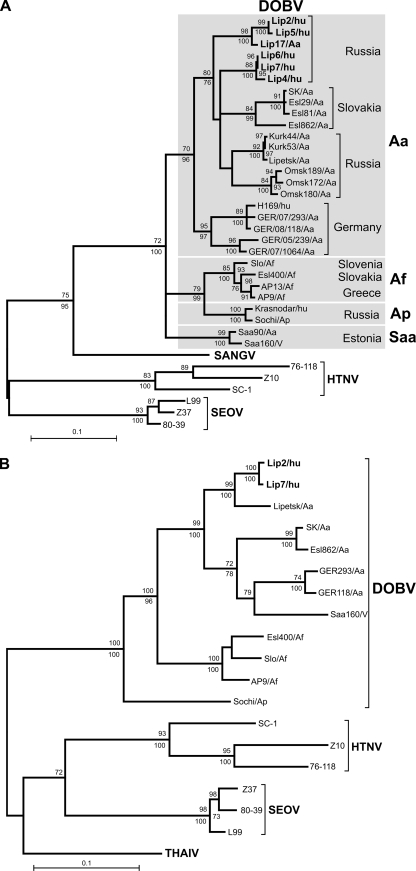
FIG. 1.
Maximum-likelihood phylogenetic trees of DOBV showing the phylogenetic placement of patient-associated sequences from the Lipetsk 2006-2007 outbreak (in boldface) on the basis of the partial S-segment nucleotide sequence (559 bp, positions 377 to 935) (A) and the partial M-segment nucleotide sequence (1,044 bp, positions 925 to 1,968) (B). In the analysis of the S segment, for which more data were available than for theMsegment, the different DOBV lineages are marked by gray boxes. The scale bar indicates an evolutionary distance of 0.1 substitutions per position in the sequence. The trees were computed with the TREE-PUZZLE package by using the Tamura-Nei evolutionary model. The values at the tree branches are the PUZZLE support values. The values below the branches are the bootstrap values of the corresponding neighborjoining tree (Tamura-Nei evolutionary model) calculated with PAUP* software from 1,000 bootstrap replicates. No differences in the tree topology were found between the maximum-likelihood and the neighbor-joining trees in the case of the M-segment tree. In the case of the S-segment trees, only minor differences that did not change the conclusions were found (trifurcations in the DOBV-Aa clade were further resolved, SK/Aa clustered with Esl29/Aa, H168/hu clustered with GER/08/118/Aa, and the DOBV-Saa clade formed an outgroup to the DOBV-Aa group in the neighborjoining tree; but this topology was not statistically supported). SANGV, Sangassou virus; THAIV, Thailand virus.