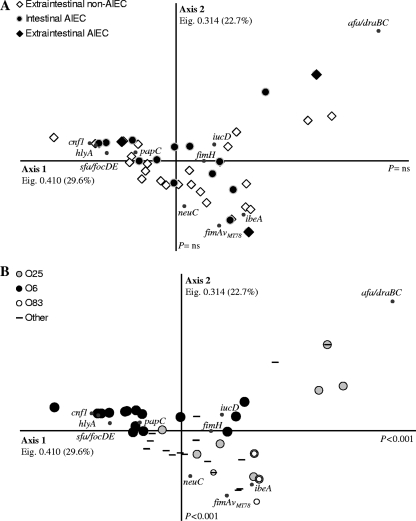
FIG. 1.
Correspondence analysis of the distribution of 10 virulence-associated genes (papC, sfa-focDE, afa-draBC, fimH, fimAvMT78, neuC, iucD, ibeA, cnf1, and hlyA) in 63 ExPEC strains and 23 intestinal AIEC strains. Eigenvalues (Eig.) and percentages of variance are provided for each axis. (A) Extraintestinal/intestinal origin of the strains and AIEC phenotype. (B) The serogroup was the sole factor that explained the segregation of the strains (only the most frequent serogroups in our collection [O6, O25, and O83] are specified). Axis 1 explains the segregation of O6 strains from the strains belonging to the O83 and O25 serogroups (P < 0.001), whereas axis 2 segregated O83 strains from the O6 and O25 serogroups (P < 0.001).