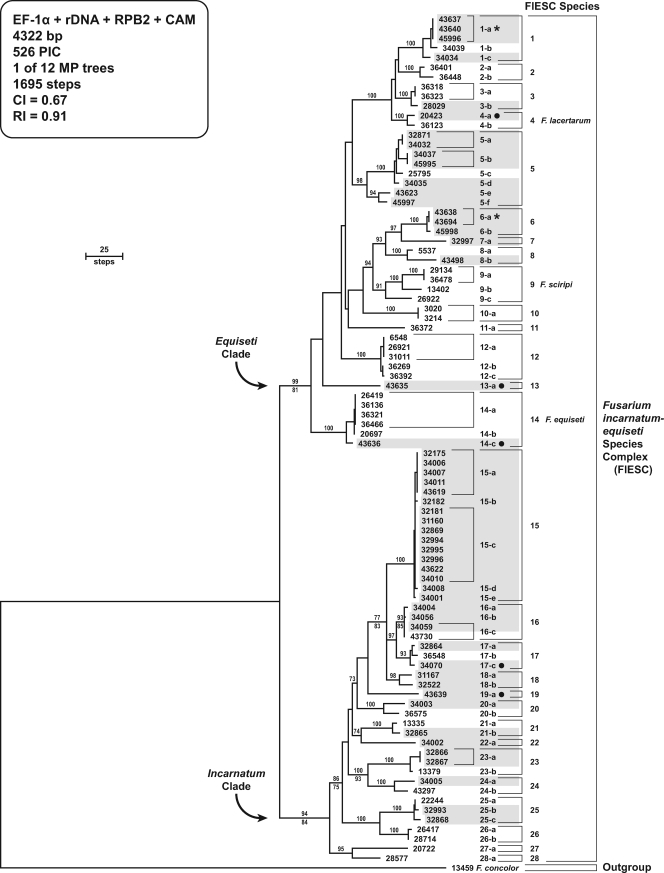
FIG. 1.
One of 12 most-parsimonious trees inferred from MP analysis of the combined four-locus data set for 88 isolates within the FIESC. The phylogram is rooted by the outgroup method using sequences of F. concolor NRRL 13459. Arabic numbers and lowercase Roman letters identify species and their multilocus STs, respectively. Human and veterinary isolates are distinguished from nonclinical isolates by shading. Veterinary isolates are distinguished from those from humans by a solid dot to the right of the NRRL number/ST. In addition, a star to the right of ST 6-a indicates that one isolate was from a veterinary source and the other from a human. Note that Latin binomials can be applied confidently to only 3 of 14 species within the Equiseti clade and to none of the species within the Incarnatum clade. MP bootstrap values based on 1,000 pseudoreplicates of the data are indicated above internodes. ML bootstrap values are indicated below internodes only when they differed by ≥5% of the MP value.