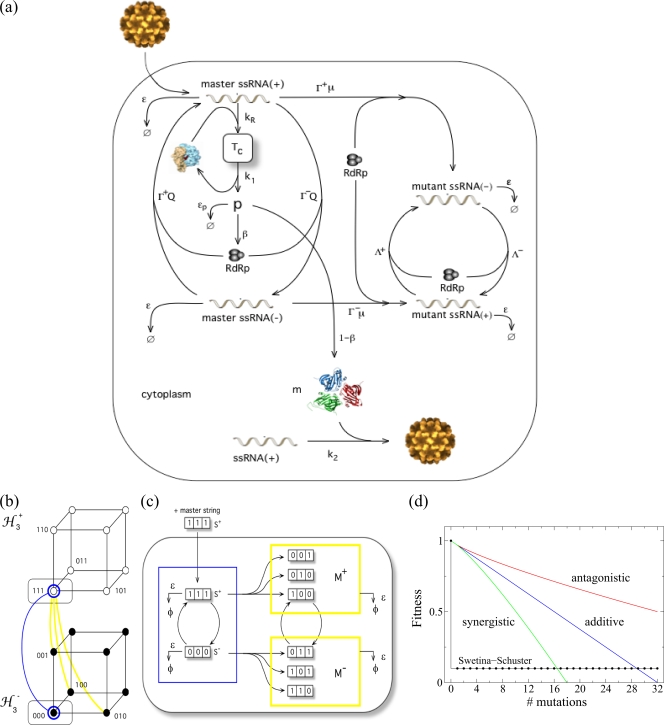
FIG. 1.
(a) Schematic representation of the virus infectious cycle. During infection, the viral particle enters into the host cell and, after uncoating, the genomic RNA (acting as mRNA) forms the translational complex, Tc, by binding with ribosomes. This directs the synthesis of the viral polyprotein, p, which is converted to both structural and nonstructural proteins at rates of 1 − β and β, respectively. The nonstructural proteins are involved in the synthesis of genomic and antigenomic RNAs. We distinguish between master and mutant genomic (+) and antigenomic (−) strands. We simulate SMR with Γ+ ≪ Γ−, where the initial genomic RNA directs the synthesis of one or very few negative copies, which are used as templates for the synthesis of new genomic strands. To model GR, we use Γ+ = Γ−, where all the synthesized strands replicate at the same rate (see Table 1 for a description of the variables and the parameters used in the model). ssRNA, single-stranded RNA; RdRp, RNA-dependent RNA polymerase. (b) Sequence spaces for a population of genomic and antigenomic binary strands of length (L = 3) bits. The consideration of genomic and antigenomic senses can be interpreted as two coupled hypercubes (ℋL±). Each node of the hypercube generates by replication of the complementary strand, SM± (blue line), or a mutant one, SM± (yellow line), in the coupled sequence space. (c) Diagram of the replication, mutation, and degradation rules implemented for the simulations. (d) Fitness landscapes analyzed in this study. The plot shows the effect of increasing numbers of mutations on the fitness of the genotype: the Swetina-Schuster (black), additive (blue), synergistic (green), and antagonistic (red) fitness landscapes. The parameters defining the landscapes shown in panel d are arbitrary.