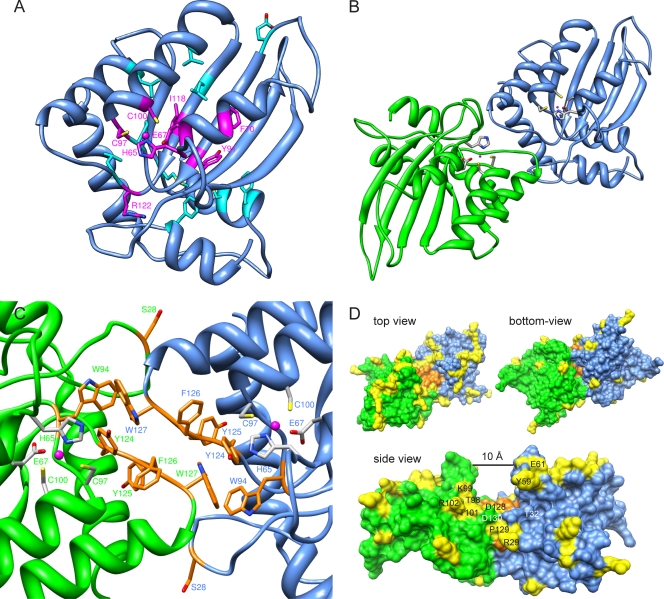
FIG. 6.
Homology dimer model of A3G. (A) A3G N-terminal domain model based on homology with the crystal structure of the A3G C terminus. Residues affecting protein steady-state level when mutated are indicated as follows. Magenta, mutants are functionally defective even after normalization for expression levels (Fig. 2C); light blue, mutants are as active as the wild type after normalization for expression levels (Phe21, Glu85, Pro96, Leu108, Leu116, Leu123, Leu138, Met152, Cys160, and Phe164). (B and C) Nitrogen, oxygen, sulfur, and zinc atoms are depicted in blue, red, yellow, and magenta, respectively. (B) Top view of the dimer model, showing two N-terminal domains of A3G (one green and the other blue) in a head-to-head complex, as modeled from the A2 tetramer structure. Zinc-binding residues (His65, Glu67, Cys97, and Cys100) are shown in gray. (C) The predicted dimer interface in detail, with residues important for A3G oligomerization and Δvif/Alu restriction shown in orange. Intersubunit contacts of the aromatic side chains allow the formation of energetically favorable π-π interactions. (D) Space-filling visualization of A3G dimer. As in panel C, the surface of the residues important for oligomerization is depicted in orange. Yellow surfaces correspond to the residues found to be under positive selection in primate A3Gs. The side view evidences the 10-Å groove formed by the two monomers at the dimer interface, as well as the residues under positive selection that are close to the Vif-binding site comprising residues 128 to 130. Thr32 and Asp130 (labeled in white) are two conserved residues that both influence recognition by HIV-1 Vif.