Abstract
Paired immunoglobulin-like type 2 receptor α (PILRα) is an inhibitory receptor expressed on both hematopoietic and nonhematopoietic cells. Its binding to a cellular ligand, CD99, depends on the presence of sialylated O-linked glycans on CD99. Glycoprotein B (gB) of herpes simplex virus type 1 (HSV-1) binds to PILRα, and this association is involved in HSV-1 infection. Here, we found that the presence of sialylated O-glycans on gB is required for gB to associate with PILRα. Furthermore, we identified two threonine residues on gB that are essential for the addition of the principal O-glycans acquired by gB and that are also essential for the binding of PILRα to gB.
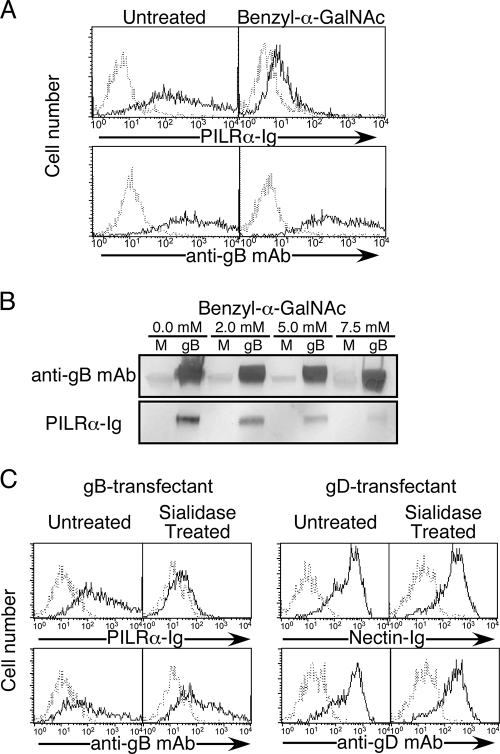
Four envelope glycoproteins, gB, gD, gH, and gL, are required for herpes simplex virus type 1 (HSV-1) to enter into host cells. Paired immunoglobulin (Ig)-like type 2 receptor α (PILRα) binds to gB and functions as an entry receptor during HSV-1 infection in concert with an interaction between gD and gD receptors (10). An X-ray structure of gB suggested that it is a class III fusogenic glycoprotein with internal fusion loops (6), and evidence that these loops can associate with lipid membranes was presented recently (5). The interaction between PILRα and gB might help the fusion loops of gB to associate with cellular membranes. However, it has remained unclear how PILRα associates with gB. PILRα also binds to CD99, which is expressed mainly on T-cell subsets (12). Specific O-glycan structures on CD99 are required for recognition of CD99 by PILRα (15). Here, we addressed whether O-glycans on gB are involved in the association between PILRα and gB. One approach was to use benzyl-α-GalNAc, which specifically blocks the extension of O-glycans through its ability to compete with GalNAc-O-Ser/Thr, a substrate for β1-3-galactosyl-transferases, which generate core 1 structures of O-glycans (8). 293T cells transfected with gB (HSV-1 strain KOS) were treated with benzyl-α-GalNAc (Sigma) at 37°C for 48 h and were then stained with PILRα-Ig (15) or anti-gB monoclonal antibody ([MAb] clone 1105; Rumbaugh-Goodwin Institute) (Fig. 1A). Recognition of gB by PILRα was abrogated almost completely by the treatment of gB transfectants with benzyl-α-GalNAc, whereas cell surface expression of gB was not affected. Because benzyl-α-GalNAc functions competitively, the weak binding of PILRα-Ig to benzyl-α-GalNAc-treated gB transfectants might have been due to an insufficient effect of benzyl-α-GalNAc on O-glycans. Benzyl-α-GalNAc did not affect the viability of cells (data not shown). Similarly, Western blot analysis showed that recognition of gB by PILRα-Ig was reduced by treatment with benzyl-α-GalNAc in a dose-dependent manner (Fig. 1B). The molecular weight of gB expressed on cells treated with benzyl-α-GalNAc was slightly lower than that of gB on untreated cells. Thus, the presence of O-glycans on gB is critical for the interaction between PILRα and gB, as it is for the interaction between PILRα and CD99 (15).
FIG. 1.
Requirement of sialylated O-glycans on gB for the interaction with PILRα. (A) 293T cells transfected with gB were treated with benzyl-α-GalNAc (10 mM) and were then stained with PILRα-Ig or anti-gB MAb (solid line). As a control, the transfectants were stained with control Ig or control MAb (dotted line). Histograms show fluorescence intensity measured in arbitrary units on a log scale (x axis) and relative cell number on a linear scale (y axis). (B) Total cell lysates of mock (M)- or gB-transfected 293T cells treated with benzyl-α-GalNAc at the indicated concentrations were separated by SDS-PAGE, followed by blotting with anti-gB antiserum (R74; see reference 2) or PILRα-Ig. (C) gB (left)- or gD (right)-transfected 293T cells were incubated in the presence or absence of sialidase (0.01 U/ml) for 3 h and were stained with PILRα-Ig (solid line), nectin-Ig (solid line), or control Ig (dotted line). Expression of gB or gD was analyzed by using anti-gB MAb (solid line), anti-gD MAb (solid line), or control MAb (dotted line). Histograms show fluorescence intensity measured in arbitrary units on a log scale (x axis) and relative cell number on a linear scale (y axis).
HSV gB is sialylated, and sialic acids on virions play an essential role in HSV-1 infection (14). Interestingly, sialic acids on O-glycans are required for recognition of CD99 by PILRα (13, 15). Therefore, we analyzed the involvement of sialic acids on gB in the interaction with PILRα. gB-transfected 293T cells treated with neuraminidase (Vibrio cholera; Roche) at 37°C for 3 h were not recognized by PILRα-Ig, whereas nontreated cells were recognized by PILRα-Ig (Fig. 1C). Neuraminidase treatment did not affect the binding of nectin-Ig to gD transfectants or the cell surface expression of gB and gD.
Four to 10% of the amino acids of PILRα are identical to Siglec (sialic acid-binding Ig-like lectin) family proteins, which recognize sialic acids on glycans (15). An arginine residue that is essential for sialic acid recognition by Siglecs is conserved in PILRα. Indeed, PILRα-Ig with this arginine residue mutated did not recognize gB or CD99 (data not shown). These results suggest that sialic acids on gB are involved in the recognition of gB by PILRα, as they are in the recognition of CD99 by PILRα. Along with the result that O glycosylation on gB is important for association with PILRα, sialylated O-glycans on gB are involved in the interaction with PILRα.
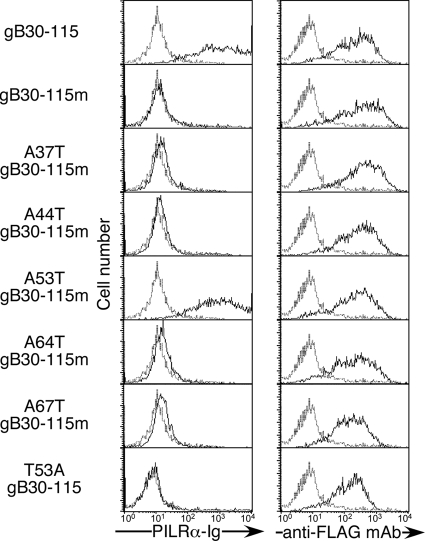
We analyzed the glycosylation sites on gB that are responsible for recognition by PILRα. Although the NetOGlyc 3.1 algorithm (www.cbs.dtu.dk/services/NetOGlyc/) is useful in predicting potential O glycosylation sites, prediction of O glycosylation sites is still imprecise. The NetOGlyc 3.1 algorithm predicted seven threonine or serine residues (threonines at 37, 44, 53, 64, 67, and 480 and serine at 487) to be potential O glycosylation sites. Of note, five threonines were located near the N terminus. In order to analyze whether this N-terminal region is involved in recognition by PILRα, we constructed a gB chimeric molecule (gB30-115) possessing a BM-40 signal sequence (amino acid residues 1 to 40), a Flag-tag, an N-terminal gB fragment from its signal peptide cleavage site (amino acid residues 30 to 115) containing the five possible O glycosylation sites, and a transmembrane region of mouse PILRα (amino acid residues 196 to 256; GenBank accession number, NM_153510) to serve as an anchor to cellular membranes. This short N-terminal fragment of gB expressed on the cell surface was stained with both anti-Flag MAb and a PILRα-Ig fusion protein similar to wild-type (WT) gB (Fig. 2). In order to identify the amino acid residues of gB that are involved in association with PILRα, we generated a series of mutations of the N-terminal gB fragment. The gB fragment, in which all possible O glycosylation sites were mutated to alanine (gB30-115m), was not recognized by PILRα-Ig, whereas cell surface expression was not affected by these mutations. A revertant that has a threonine at amino acid residue 53 (A53T gB30-115m) was recognized by PILRα-Ig. In contrast, a WT N-terminal gB fragment in which only threonine 53 (T53) was mutated to alanine (T53A gB30-115) was not recognized by PILRα-Ig. Furthermore, the binding of PILRα-Ig to the A53T gB30-115m revertant was abrogated by sialidase or benzyl-α-GalNAc treatment (data not shown). Therefore, T53 is the only threonine within residues 30 to 115 of gB whose O glycosylation is required for the association of gB with PILRα.
FIG. 2.
Mutational analyses of O glycosylation sites in the N terminus domain of gB. Flag-tagged N terminus fragments of gB (amino acid residues 30 to 115) containing five potential O glycosylation sites or point mutations of these possible O glycosylation sites were transfected into 293T cells. The transfectants were stained with control Ig (dotted line) or PILRα-Ig (solid line). Expression of the N terminus domain of gB was analyzed by staining with anti-Flag MAb (solid line) or control MAb (dotted line). Histograms show fluorescence intensity measured in arbitrary units on a log scale (x axis) and relative cell number on a linear scale (y axis).
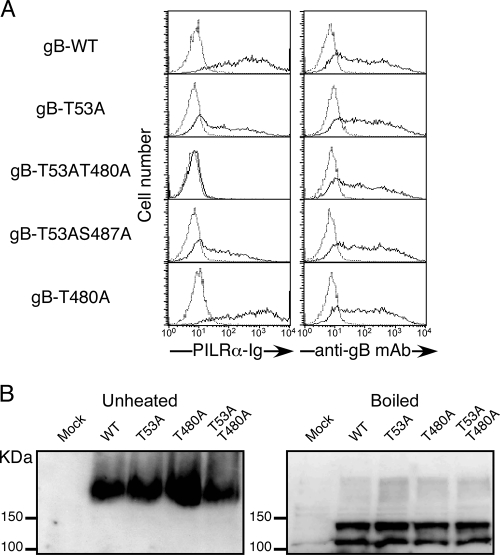
We generated full-length gB in which T53 was mutated to alanine (T53A gB). The single point mutation at T53 partially affected the recognition of full-length gB by PILRα-Ig, whereas cell surface expression of gB was not affected (Fig. 3A). This finding suggests that T53 is a dominant O glycosylation site on gB, which is involved in interactions with PILRα, although additional potential O glycosylation sites other than those near the N terminus might also be involved. Interestingly, gB with a mutation at threonine 480 (T480) in addition to T53 (T53AT480A) was not recognized by PILRα, whereas, similar to T53A gB, gB with an additional mutation at serine 487 (T53AS487A) was recognized by PILRα-Ig. gB with a mutation at T480 alone (T480A) was recognized by PILRα, as was WT gB. None of these mutations affected the cell surface expression of gB. Similar results were obtained using several other cell lines, such as COS cells (data not shown). These data suggest that two O glycosylation sites of gB, T53 and T480, are involved in the association with PILRα.
FIG. 3.
Mutational analyses of O glycosylation sites of full-length gB. (A) 293T cells were transfected with various mutated gBs, and the transfectants were stained with control Ig (dotted line) or PILRα-Ig (solid line). Expression of gB was analyzed by staining with anti-gB MAb (solid line) or control MAb (dotted line). The histograms show fluorescence intensity measured in arbitrary units on a log scale (x axis) and relative cell number on a linear scale (y axis). (B) Membrane proteins prepared from COS-7 cells transfected with WT gB and mutated gBs were boiled or left unheated in SDS sample buffer in reducing or nonreducing conditions, respectively. Samples were separated by SDS-PAGE, followed by blotting with anti-gB MAb.
Both T53 and T480 are located in a proline-rich region, which may be important for protein folding (16). It has been reported that functional gB forms oligomers (1, 3, 6). Therefore, we analyzed whether the point mutations of gB affected oligomer formation. The oligomeric structure of gB is resistant in sodium dodecyl sulfate (SDS) sample buffer but is denatured by boiling (9). As shown in Fig. 3B, there was no difference in SDS resistance between WT gB and the mutated gBs. This suggests that point mutations of the O glycosylation sites at T53 and T480 of gB did not greatly affect the physical characteristics of gB. Moreover, there was no difference in the molecular weight between WT and mutated gB or in cell surface expression. Because the molecular weight of gB is relatively high and gB has several N glycosylation sites, mutations of one or two O glycosylation sites alone did not affect the total molecular weight of gB. However, the molecular weight of gB expressed in benzyl-α-GalNAc-treated cells was slightly lower than that of gB expressed in nontreated cells (Fig. 1B). Because benzyl-α-GalNAc treatment inhibits synthesis of all the O-glycans on gB, other O glycosylation sites on gB might exist. However, it is noteworthy that only O glycosylation sites at T53 and T480 are involved in association with PILRα.
Although mutations at T53 and T480 of gB completely abrogated recognition by PILRα, there is no direct evidence to suggest that these two residues are O glycosylated. In order to analyze O glycosylation on gB, we employed a novel method to label O-linked glycans, using Click-iT O-GalNAz metabolic glycoprotein-labeling reagent (azido-GalNAc) (Invitrogen). Because O-linked glycans generally possess peptide-proximal GalNAc residues (7), we cultured cells transfected with WT gB or mutated gB for 3 days in the presence of azido-GalNAc, which is metabolically incorporated into O-linked glycoproteins (4). gBs were immunoprecipitated with anti-gB MAb, and the azido-GalNAc incorporated into gB was treated with phosphine-Flag, which specifically reacts with the azido-GalNAc (11), followed by detection with anti-Flag MAb by Western blotting. WT gB, T53A-mutated gB, and T480A-mutated gB were blotted with anti-Flag MAb, whereas T53AT480A gB was only weakly blotted with anti-Flag MAb (Fig. 4). In contrast, there was no significant difference in the total amount of gB expressed. This result suggests that T53 and T480 of gB are O glycosylated. However, weak detection of O-glycans on the T53AT480A gB suggest the presence of O glycosylation sites other than T53 and T480 on gB.
FIG. 4.
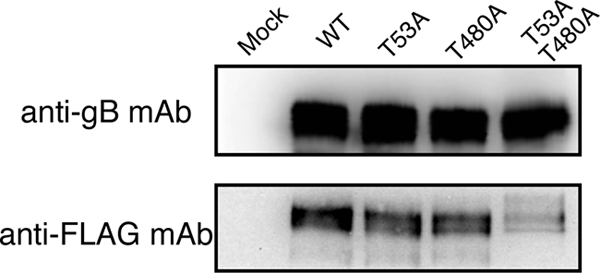
Analysis of O-glycans on WT and mutated gB. O-glycans on gB expressed on 293T cells were metabolically labeled with Ac4GalNAz and were then immunoprecipitated with anti-gB MAb. The labeled O-glycans in immunoprecipitates were modified with phosphine-Flag and were then analyzed by Western blotting. The labeled O-glycans and total amount of gB were detected by anti-Flag or anti-gB MAb, respectively.
PILRα specifically associates with HSV-1 gB (10), but not with other HSV-1 glycoproteins, although some other envelope proteins are known to be O glycosylated. Recently, it was shown that insertion mutations in gB could reduce the binding of gB to PILRα, suggesting that the conformation of gB is also involved in the interaction (2). Therefore, PILRα does not associate with glycans alone and seems to recognize both protein structure and O-glycans (13, 15), which may be a reason that PILRα specifically associates with gB. It is interesting that both T53 and T480 are involved in the interaction with PILRα, because these two residues are widely separated on the polypeptide chain. Because PILRα bound to gB by Western blotting, PILRα might recognize linear epitopes in gB; therefore, PILRα might bind to the two sites in gB independently. Alternatively, elements of higher-order structure retained in the unreduced samples examined by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 1B) could have been necessary for the binding of PILRα-Ig to the blots. Thus, the binding of PILRα might depend upon the close proximity of the O-glycans attached to T53 and T480 in the trimeric conformation of gB. Determination of the structure of gB associated with PILRα will facilitate understanding the mechanism of membrane fusion during HSV-1 infection.
Acknowledgments
We thank K. Maenaka for discussions and R. Hirohata and M. Matsumoto for technical assistance.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture, Japan (Y.K. and H.A.), and in part by grants from the Takeda Science Foundation (Y.K. and H.A.) and NIH grants R01AI068129 (L.L.L.), R37AI036293 (P.G.S.), and R01CA021776 (P.G.S). L.L.L. is an American Cancer Society Research Professor.
Footnotes
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Claesson-Welsh, L., and P. G. Spear. 1986. Oligomerization of herpes simplex virus glycoprotein B. J. Virol. 60:803-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan, Q., E. Lin, T. Satoh, H. Arase, and P. G. Spear. 2009. Differential effects on cell fusion activity of mutations in herpes simplex virus 1 glycoprotein B (gB) dependent on whether a gD receptor or a gB receptor is overexpressed. J. Virol. 83:7384-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Handler, C. G., R. J. Eisenberg, and G. H. Cohen. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067-6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hang, H. C., C. Yu, D. L. Kato, and C. R. Bertozzi. 2003. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. USA 100:14846-14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannah, B. P., T. M. Cairns, F. C. Bender, J. C. Whitbeck, H. Lou, R. J. Eisenberg, and G. H. Cohen. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 7.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153-164. [DOI] [PubMed] [Google Scholar]
- 8.Kuan, S. F., J. C. Byrd, C. Basbaum, and Y. S. Kim. 1989. Inhibition of mucin glycosylation by aryl-N-acetyl-α-galactosaminides in human colon cancer cells. J. Biol. Chem. 264:19271-19277. [PubMed] [Google Scholar]
- 9.Lin, E., and P. G. Spear. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. USA 104:13140-13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoh, T., J. Arii, T. Suenaga, J. Wang, A. Kogure, J. Uehori, N. Arase, I. Shiratori, S. Tanaka, Y. Kawaguchi, P. G. Spear, L. L. Lanier, and H. Arase. 2008. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxon, E., and C. R. Bertozzi. 2000. Cell surface engineering by a modified Staudinger reaction. Science 287:2007-2010. [DOI] [PubMed] [Google Scholar]
- 12.Shiratori, I., K. Ogasawara, T. Saito, L. L. Lanier, and H. Arase. 2004. Activation of natural killer cells and dendritic cells upon recognition of a novel CD99-like ligand by paired immunoglobulin-like type 2 receptor. J. Exp. Med. 199:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabata, S., K. Kuroki, J. Wang, M. Kajikawa, I. Shiratori, D. Kohda, H. Arase, and K. Maenaka. 2008. Biophysical characterization of O-glycosylated CD99 recognition by paired Ig-like type 2 receptors. J. Biol. Chem. 283:8893-8901. [DOI] [PubMed] [Google Scholar]
- 14.Teuton, J. R., and C. R. Brandt. 2007. Sialic acid on herpes simplex virus type 1 envelope glycoproteins is required for efficient infection of cells. J. Virol. 81:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, J., I. Shiratori, T. Satoh, L. L. Lanier, and H. Arase. 2008. An essential role of sialylated O-linked sugar chains in the recognition of mouse CD99 by paired Ig-like type 2 receptor (PILR). J. Immunol. 180:1686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]