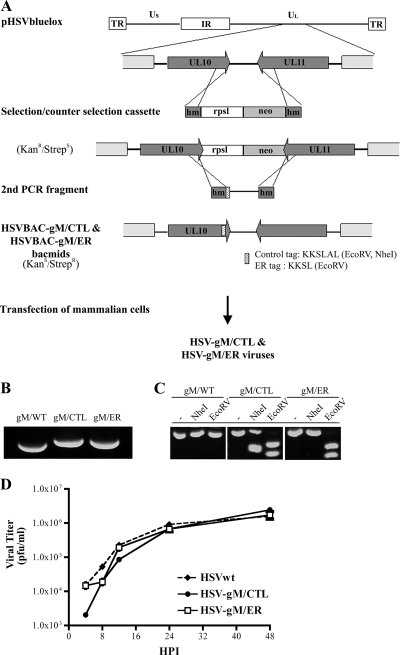
FIG. 1.
Construction and generation of recombinant virus. (A) Schematic diagram of the selection and counterselection approach to introduce gM/KKSL (ER retention signal) or gM/KKSLAL (inactive signal) into the viral genome of HSV-1 strain 17+ present in the pHSV1(17+)bluelox bacmid. Shown at the top is the viral genome map with the unique long (Ul) and unique short (Us) regions, the internal repeats (IR), and the terminal repeats (TR) and the enlargement of the UL10 (gM) and UL11 gene regions. The mutagenesis involved the synthesis by PCR of a selection and counterselection cassette encoding a selectable (neo; light-gray box) and a counterselectable (RpsL; white box) marker flanked by HSV-1 sequences (dark-gray box). Following the insertion of this cassette into the pHSV1(17+)bluelox backbone by homologous recombination, a second PCR fragment covering the end region of the tagged gM and containing the control or ER retention tag was amplified from pHSV1(17+)bluelox and used to subsequently remove the selection cassette, once again by homologous recombination. The stippled box represents the tag sequences into which restriction sites were introduced for diagnostic purposes. hm, homologous sequences; RpsL, streptomycin sensitivity gene; neo, kanamycin resistance gene. (B and C) The recombinant BACs (HSVBAC-gM/CTL and HSVBAC-gM/ER) were finally transfected into 143B cells to reconstitute recombinant viruses. Analyses of these viruses by PCR (B) and restriction enzyme digestion (C) were performed to verify the mutagenesis (see the text for details). (D) A single-step growth kinetics assay of wild-type HSV (HSVwt), HSV-gM/CTL, and HSV-gM/ER was performed. 143B cells were infected at an MOI of 5, and the supernatant was harvested at the indicated time and concentrated by centrifugation. The yields of released infectious virus were then determined by plaque assays on Vero cells. All values are the means of three independent experiments, and the small error bars depict the standard deviations of the means.