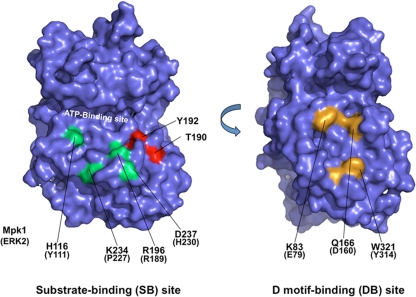
FIG. 1.
Positions of substrate-binding site mutations and D-motif-binding site mutations within the predicted structure of Mpk1. The structure was modeled against known MAPK structures using the SWISS-MODEL program (http://swissmodel.expasy.org/SWISS-MODEL.html). The two images are rotated 180° with respect to each other. Mutated substrate-binding site residues are indicated in green, and mutated D-motif-binding site residues are indicated in yellow. Residues phosphorylated in the activation loop (T190 and Y192) are indicated in red.