Figure 1.
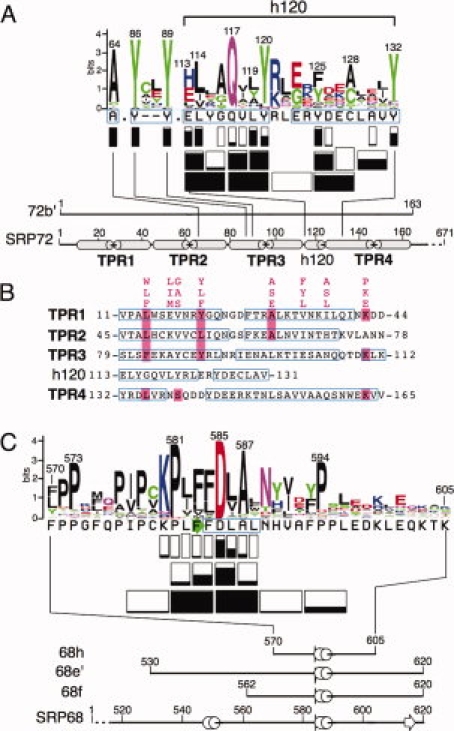
Properties of the interface between human SRP72 and SRP68. (A) The protein sequence logo of the 72b′ region was prepared using the interface at http://weblogo.berkeley.edu/logo.cgi. The height of each symbol is proportional to its frequency in the alignment of representative SRP72 sequences obtained as described in “Materials and Methods” section. Rectangles below each sequence indicate how many residues were altered. Negative effects of the ability to form complexes are shown by the extent of black filling and are listed in Table I. Not considered are the results obtained from competition experiments with A117–118 Q117A, and V118A [Fig. 4(D)]. Below the logo, amino acid positions are labeled in 20-residue increments according to full-length human SRP72. The termini of fragment 72b′ are numbered. Alpha helices, predicted from alignments of representative eukaryotic sequences (available at http://rnp.uthct.edu/rnp/SRPDB/srpprotein.html) are marked as gray cylinders. The positions of four tetratricopeptide repeats (TPR1 to TPR4) as well as a small predicted alpha-helix located between TPR3 and TPR4 (h120) are indicated. (B) Alignment of the four N-terminal TPR-like regions of human SRP72 and the h120 insertion between TPR3 and TPR4. The TPR consensus sequence24 and matching SRP72 residues are highlighted in pink. Amino acid residues are numbered according to full-length human SRP72.23 Regions predicted to be alpha-helical as determined by PSIPRED25 are shown in blue frames. (C) Protein logo of the 68h region with effects of mutant polypeptides depicted as in panel A. The termini of fragments 68e′, 68f, and 68h are numbered according to full-length human SRP68. Alpha helices and a beta turn, predicted from alignments of representative SRP68 sequences, are marked in blue and as a green arrow, respectively.
