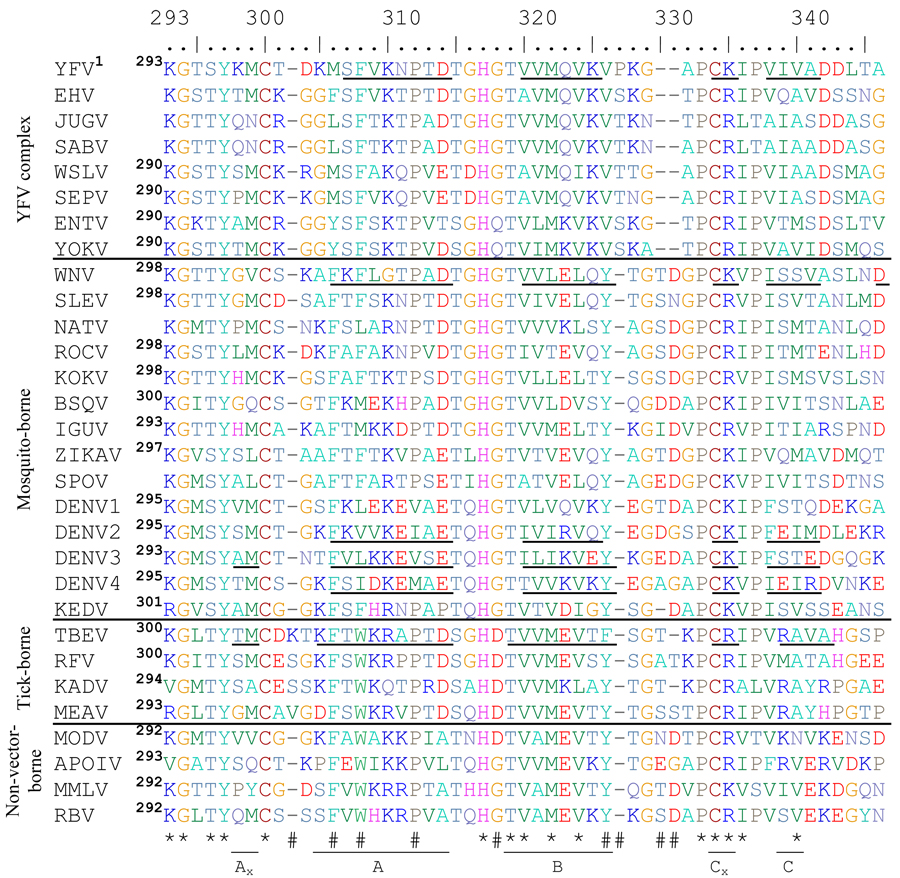
Table 2.
Alignment of flavivirus ED3 proteins from mosquito-, tick- and non-vector-borne flaviviruses. Asterisks (*) indicate highly conserved residues and (#) hashes indicate residues with group-specific differences. Beta strands are indicated with underlined residues and are labeled underneath using TBEV(Rey et al., 1995) nomenclature. The number of the first amino acid of ED3 for each flavivirus is shown in superscript to the right of the name of each flavivirus. Those without available full length E genes sequences are left blank. Biochemically similar amino acids are the same color to allow easier understanding of the alignment.
 |
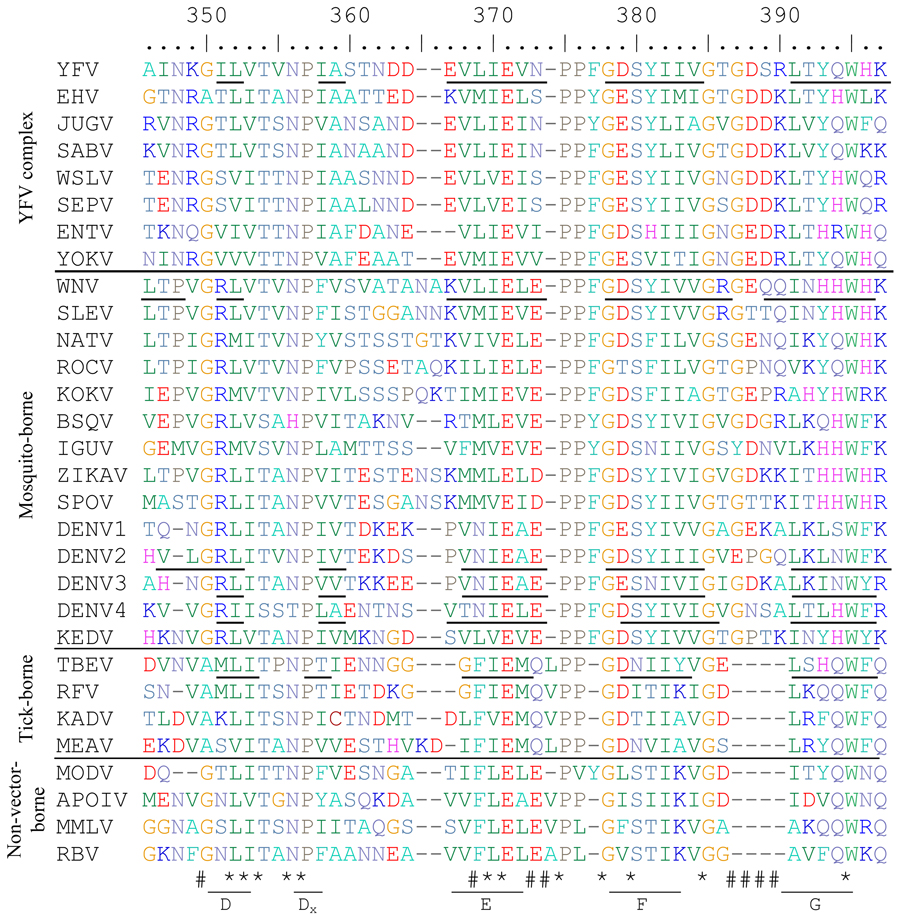 |
ICTV abbreviations used are as follows: Yellow fever virus=YFV, Edge Hill virus=EHV, Jugra virus=JUGV, Saboya virus=SABV, Wesselsbron virus=WSLV, Sepik virus=SEPV, Entebbe bat virus=ENTV, Yokose virus=YOKV, West Nile virus=WNV, St Louis encephalitis virus=SLEV, Ntaya virus=NATV, Rocio virus=ROCV, Kokobera virus=KOKV, Bussuquara virus=BSQV, Iguape virus=IGUV, Zika virus=ZIKAV, Spondweni virus=SPOV, dengue virus=DENV, Kedougou virus=KEDV, tick-borne encephalitis virus=TBEV, Royal Farm virus=RFV, Kadam virus=KADV, Meaban virus=MEAV, Modoc virus=MODV, Apoi virus=APOIV, Montana myotis leukoencephalitis virus=MMLV, Rio Bravo virus=RBV
