Abstract
Caenorhabditis elegans fem-3 binding factor (FBF) is a founding member of the PUMILIO/FBF (PUF) family of mRNA regulatory proteins. It regulates multiple mRNAs critical for stem cell maintenance and germline development. Here, we report crystal structures of FBF in complex with 6 different 9-nt RNA sequences, including elements from 4 natural mRNAs. These structures reveal that FBF binds to conserved bases at positions 1–3 and 7–8. The key specificity determinant of FBF vs. other PUF proteins lies in positions 4–6. In FBF/RNA complexes, these bases stack directly with one another and turn away from the RNA-binding surface. A short region of FBF is sufficient to impart its unique specificity and lies directly opposite the flipped bases. We suggest that this region imposes a flattened curvature on the protein; hence, the requirement for the additional nucleotide. The principles of FBF/RNA recognition suggest a general mechanism by which PUF proteins recognize distinct families of RNAs yet exploit very nearly identical atomic contacts in doing so.
Keywords: crystal structure, RNA, translational regulation, fem-3 binding factor, base flipping
mRNA regulation permeates biology. Elements in 3′-UTRs are recognized by regulatory proteins that activate, repress, stabilize, and destroy target mRNAs. Their specificity defines which mRNAs are regulated, and so determines biological outcomes. PUMILIO/fem-3 binding factor (FBF) proteins (1, 2), or PUF proteins, are exemplary and regulate batteries of mRNAs to control stem cell maintenance and memory (1, 3–5). Understanding the structural basis of their RNA-binding specificities is a critical goal.
The PUF protein family includes members throughout eukaryotes. To date, PUF proteins invariably bind to elements in 3′-UTRs (1). In most instances, the PUF/RNA complexes decrease expression of the mRNA, although activation also can occur (6, 7). PUF proteins contain a conserved RNA-binding domain, known as the Pumilio-homology domain or PUF domain. This domain comprises 8 PUF repeats, flanked by pseudorepeats (8–11). The PUF domain is sufficient to bind target mRNAs (8, 10, 12), recruit partner proteins (13–16), and regulate mRNAs in vivo (2, 17).
A typical PUF repeat contains 3 α-helices, and the 8 repeats in a single protein pack to form a crescent shape (16, 18). In human PUMILIO1 (PUM1) bound to a nanos response element (NRE) RNA, each PUF repeat interacts with a single RNA base along an extended concave surface (19). In a typical PUF repeat, the second α-helix provides 3 conserved amino acids that interact with an RNA base. Two residues contact the edge of the base via hydrogen bonding or van der Waals interactions, and a third often is sandwiched between 2 bases to form stacking interactions. The identities of the residues contacting the edge of the bases play a key role in selecting the RNA base (19–22).
The Caenorhabditis elegans PUF proteins, FBF-1 and FBF-2, provide a special opportunity to dissect the structural basis of PUF–RNA interactions in a biological context. FBF-1 and FBF-2 are 91% identical in amino acid sequence, have similar RNA-binding specificity, and overlap functionally (8, 23–27); they are referred to collectively as FBF. Eight mRNA targets of FBF have been identified using a combination of in vitro binding, physical association in worm extracts, immunocytochemistry, and genetics (5, 8, 23–27). FBF is required for maintenance of germline stem cells, a conserved function of PUF proteins (1). FBF maintains stem cells by repressing the expression of differentiation regulators, including gld-1, fog-1, lip-1, and mpk-1 (28). In addition, FBF regulates the switch from spermatogenesis to oogenesis by repressing fem-3 mRNA (8). The regulatory elements found in the different mRNAs are related, but distinct. PUF proteins coimmunoprecipitate with many mRNAs in yeast, fly, and human extracts (4, 29, 30). Although the biological relevance of all the interactions is not yet clear, the conclusion that a single PUF protein associates with a large number of functionally related mRNAs is inescapable.
The biochemical basis of FBF's specificity for RNA has been examined in depth and provides a foundation for structural analysis. FBF binds a 9-nt sequence, despite having only 8 PUF repeats (20). The core FBF recognition sequence begins with a UGU triplet, as do all validated PUF target sequences. The optimal binding site is 5′-UGURNNAUA-3′ (R, purine; N, any base; see Results) (28). PUF-8, a close relative in C. elegans to PUMILIO, recognizes a core 8-nt sequence (20), 5′-UGUANAUA-3′ (4, 30–32), the same as that seen by PUM1 (30). Biochemistry and genetics identified the fifth position of the FBF recognition sequence as an inserted base relative to the PUF-8 (20). It has been proposed that a structural distortion in the central region of the protein imposes a requirement for a “spacer” nucleotide (20). This “extra” base is an identity determinant that enables the protein to bind and regulate cognate mRNAs uniquely.
We sought to understand how FBF specifically recognizes its 9-nt mRNA targets despite having 8 repeats. To do so, we determined crystal structures of the RNA-binding domain of FBF-2 and 6 different RNA sequences, including 4 natural target sequences. The data not only reveal the basis of FBF specificity but lead to general models of PUF protein RNA specificity and their evolution.
Results
Crystal Structures of FBF-2 in Complex with 9-nt RNA Target Sequences.
A crystal structure of the RNA-binding domain of FBF-2 in complex with a 9-nt consensus FBF binding element (FBE), 5′-UGUACUAUA-3′ (20, 28), was determined by single wavelength anomalous diffraction (SAD) and refined to a resolution of 2.2 Å. The initial experimental electron density map revealed density for all 9 RNA bases [supporting information (SI) Fig. S1]. As a convention, we designate the 5′-UGU as positions 1–3 in the recognition sequences.
We subsequently determined the crystal structures of FBF-2 in complex with 9-nt sequences from the 3′-UTRs of natural mRNA targets: the gld-1 FBEa and FBEb mediate repression of gld-1 mRNA to control stem cells (23); the fog-1 FBEa triggers repression to permit oogenesis (27); and the fem-3 point mutation element (PME) mediates repression to permit the switch from spermatogenesis to oogenesis (27). In addition, we determined the structure of a complex with a mutation in the gld-1 FBEa, G4A. The protein structures in the different complexes are very similar (rmsd <0.6 Å). We use the structure of FBF-2 bound to the gld-1 FBEa RNA, the highest affinity natural target sequence, to present general features of FBF-2 RNA recognition.
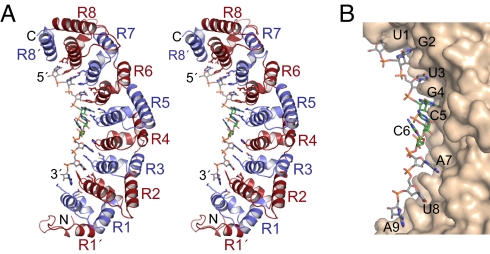
Overall, the structure of the RNA-binding domain of FBF-2 is similar to that of other PUF proteins, with 8 α-helical PUF repeats (R1–R8) and flanking regions at the N- and C-termini [R1′ and R8′, including Csp1a and Csp2 (8), respectively; Fig. 1A] and the 9-nt RNA target sequence bound to the concave surface (Fig. 1B). FBF-2 contacts mainly the bases (Fig. S2), leaving ribose and phosphate groups facing the solvent.
Fig. 1.
Crystal structure of FBF-2 in complex with gld-1 FBEa RNA. (A) Stereo-view of FBF-2 in complex with gld-1 FBEa RNA. Repeats are colored alternately red and blue. Side chains that interact with RNA are shown. The RNA is colored by atom type (gray, carbon; red, oxygen; blue, nitrogen; orange, phosphorus; yellow, sulfur). Bases 4–6 are shown with green carbon atoms. (B) Surface representation of FBF-2 in complex with gld-1 FBEa RNA. The figures were prepared with PyMol (44).
The lengths of PUF repeats vary in FBF from 38–56 residues. In contrast, human and fly PUMILIO have uniform lengths (36 aa) (16, 18). The additional residues in FBF are incorporated into a longer helix (third helix in repeat 5) and longer loop regions between the second and third helices in repeats 2, 5, and 8 and between repeats 7 and 8 (Fig. S3).
Recognition of Conserved RNA Regions by FBF-2.
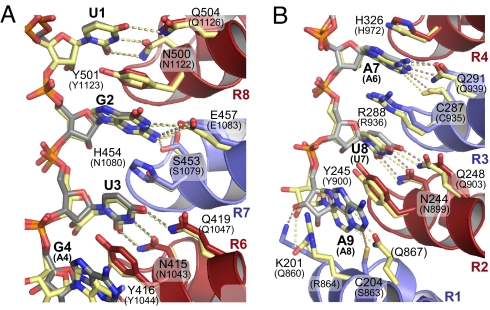
The architecture of FBF-2/RNA complexes consists of conserved amino acid–nucleotide interactions at the 2 ends of the RNA sequence, which bracket a unique central region in which the bases flip away from the protein. Here, we focus initially on the conserved bracketing interactions. The FBF-2 RNA recognition sequence contains 2 highly conserved regions, the 5′-UGU sequence commonly found in PUF protein target sequences and a downstream A7–U8 sequence. The regions are recognized similarly by FBF-2 and PUM1 (Fig. 2 and Fig. S2).
Fig. 2.
Recognition of conserved RNA regions by FBF-2. (A) Conservation of PUF protein recognition of the 5′-conserved UGU sequence. Superposition of repeats 6–8 in the structures of FBF-2 in complex with gld-1 FBEa RNA and PUM1 with NRE RNA. FBF-2 is depicted as in Fig. 1A. The NRE RNA and RNA-interacting side chains of PUM1 are colored yellow. Dashed lines indicate interacting atoms (gray, FBF; yellow, PUM1). Corresponding CA atoms in repeats 6–8 of FBF-2 and PUM1 were aligned (rmsd of 1.41 Å over 105 CA atoms). The RNA-interacting side chains of PUM1 are labeled in parentheses. (B) Conservation of PUF protein recognition at the 3′-end. Corresponding CA atoms in repeats 1–3 of FBF-2 and PUM1 were aligned (rmsd of 1.70 Å over 99 CA atoms).
The fourth position, a purine in all validated PUF sites, is recognized differently by FBF-2 whether it is G or A. In both cases, Y416 from repeat 6 stacks with the base (Figs. 2A and 3) but different side chain and main chain atoms contact the edge of the bases (Fig. S2). The ninth position is not well conserved in natural targets, but RNA selections reveal a preference for A (28). In the FBF-2 structures, the ninth base stacks with Y245 of repeat 2 and is recognized by C204 or E208 from repeat 1, depending on the identity of the base (Fig. S4).
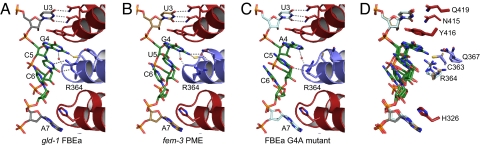
Fig. 3.
Three classes of RNA conformation in FBF-2 target RNAs. Interaction of FBF-2 repeats 4–6 with gld-1 FBEa (A), fem-3 PME (B), and gld-1 FBEa G4A mutant (C) RNAs are shown. Dashed lines indicate interacting atoms. Water molecules that mediate interactions are shown as red spheres. (D) Superposition of the RNA-interacting side chains of FBF-2 repeats 4–6 and gld-1 FBEa (gray), fem-3 PME (tan), and gld-1 FBEa G4A mutant (pale cyan) RNAs (rmsd of 0.15 Å over 130 CA atoms between PME and FBEa and rmsd of 0.28 Å over 130 CA atoms between PME and FBEa G4A mutant).
Coordinated Stacking and Base Flipping Is a Key Determinant.
A critical determinant of FBF's RNA-binding specificity lies in the central region of the FBF/RNA complex. There, bases 5 and 6 stack with each other and base 4 and turn away from the RNA-binding surface (Figs. 1B and 3). R364 in a typical repeat would form stacking interactions between the fourth and fifth RNA bases; however, instead, that side chain is moved aside and forms different contacts with the fifth base, depending on the identity of the fourth and fifth bases. The structure explains why FBF has little specificity for positions 5 and 6 (28): Interactions between R364 and the fifth base are plastic, and the sixth base does not interact at all. The flexibility of the FBF/RNA interaction includes modest sequence-dependent alterations in the RNA structure from bases 4–6. RNA conformations in this central region can be grouped into 3 different classes, depending on the identities of bases 4–6 (Fig. 3 and Fig. S5). The conformation of the R364 side chain differs for each class. All sugar puckers are in the C3′-endo conformation, and classification of the RNA structural suite is unchanged (33, 34).
Structural Basis of Affinities for Different RNAs.
In vivo, FBF regulates mRNA target sequences that vary at positions 4–6 and 9. RNA selection experiments show variability at these positions as well (28). To probe RNA sequence preferences of FBF-2, we quantified RNA-binding affinities of FBF-2 for the 9-nt RNAs used in our structural studies (Table 1). We analyzed binding to 2 additional natural target RNAs, ced-4 FBEa and mpk-1 FBEb, and 2 RNAs in which the ninth position of fem-3 PME was changed to either A or C. We confirmed weak binding to an 8-nt NRE RNA.
Table 1.
RNA-binding analyses of FBF-2
| RNA | RNA sequence* | Kd (nM) | Krel† |
|---|---|---|---|
| 123456789 | |||
| gld-1 FBEa | UGUGCCAUA | 33.2 ± 1.5 | 1.0 |
| gld-1 FBEa G4A | UGUACCAUA | 39.4 ± 1.3 | 1.2 |
| FBE | UGUACUAUA | 45.8 ± 3.2 | 1.4‡ |
| fem-3 PME | UGUGUCAUU | 52.9 ± 2.4 | 1.6‡ |
| PME_U9A | UGUGUCAUA | 56.7 ± 3.4 | 1.7‡ |
| PME_U9C | UGUGUCAUC | 81.3 ± 3.5 | 2.4‡ |
| ced-4 FBEa | UGUACAAUA | 73.6 ± 1.7 | 2.2‡ |
| mpk-1 FBEb | UGUAAUAUA | 81.7 ± 2.8 | 2.5‡ |
| fog-1 FBEa | UGUAAAAUC | 96.9 ± 3.4 | 2.9‡ |
| gld-1 FBEb | UGUGUUAUC | 127 ± 6.4 | 3.8‡ |
| NRE | UGUAUAUA | >1,000§ | >30‡ |
*Numbers indicate positions in the core recognition sequence. Bases that differ from the sequence of gld-1 FBEa are shown in boldface.
†Krel reports the affinity of FBF-2 for the RNA sequence relative to the affinity of FBF-2 for gld-1 FBEa RNA. Double daggers (‡) denote statistically significant differences (P < 0.05).
§Saturable binding could not be achieved at the highest protein concentration; therefore, we report a lower limit for the Kd.
The binding studies lead to 5 main conclusions with respect to positions 4, 5, 6, and 9:
Range of affinities. The Kds of natural target RNA sequences tested, bearing substitutions at positions 4–6 and 9, range from 30–130 nM. The tightest binding natural target RNA (gld-1 FBEa) binds 4-fold more tightly than the weakest binding natural target RNA (gld-1 FBEb).
Position 4: Adenosine and guanine bind equally. An RNA with a G4A mutation binds as well as G4 (compare gld-1 FBEa and gld-1 FBEa mutant; P = 0.07). Prior selection experiments and mutational analysis show a preference for A (28). Natural target RNAs often possess a G at this position, and the structures suggest that either base is recognized, mainly through water-mediated contacts.
Position 5: Modest preference. C5 binds ≈2-fold tighter than U5 (compare gld-1 FBEa with fem-3 PME_U9A; P = 0.04) or A5 (compare FBE with mpk-1 FBEb; P = 0.001), consistent with the fact that R364 makes direct hydrogen-bonding contacts with C5 but not with U5 or A5. Natural target RNA sequences, selection experiments, and mutational analysis corroborate that FBF-2 allows U, C, or A at bases 5 and 6 (20, 28).
Position 6: Flexibility, C = U > A. C6 and U6 bind equivalently well (compare fem-3 PME_U9C vs. gld-1 FBEb; P = 0.06 or gld-1 FBEa G4A mutant vs. FBE; P = 0.14). Both C6 and U6 bind ≈2-fold better than A6 (compare gld-1 FBEa with ced-4 FBEa; P = 0.02 or FBE with ced-4 FBEa; P = 0.02). These differences likely arise through effects on base stacking, because base 6 does not contact the protein. Natural target RNA sequences, selection experiments, and mutational analysis indicate variability at this position (20, 28).
Position 9: Modest preferences via varying contacts. Modest preferences at base 9 indicate A = U > C (compare PME, PME_U9A, and PME_U9C). In the crystal structures, the position of the ninth base is somewhat different depending on the identity of the base (Fig. S4). Natural target mRNAs can contain any nucleotide at this location. Mutational analysis is consistent with the pattern in our binding data; however, in selection experiments, A is preferred (28).
Combinations of base changes affect binding predictably and suggest that the effects of base variations, although small, together influence relative binding affinity. For example, the weakest binding natural RNA targets in our study are fog-1 FBEa and gld-1 FBEb, which bear suboptimal sequences A5–A6/C9 and U5/C9, respectively. In contrast, gld-1 FBEa, which carries optimal bases at positions 5, 6, and 9, binds 4-fold more tightly than gld-1 FBEb, which has suboptimal bases at these positions.
Curvature Imposed by a Central Region Yields Specificity.
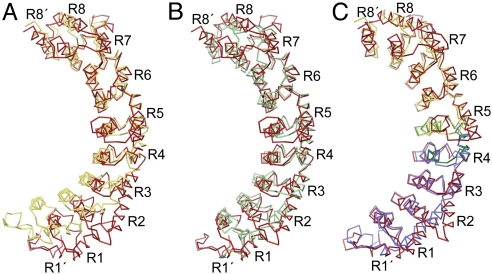
FBF-2 has a distinctive curvature that may impose the requirement for its extra base relative to 8-nt binding PUF proteins, such as PUM1. The concave RNA-binding surface of FBF-2 is flatter than in PUM1, and the α-helices that line the surface are twisted (Fig. 4A, compare the position of the RNA-binding helices of FBF-2 and PUM1 in repeats 1–3), creating an elongated RNA-binding surface. The degree of curvature is similar to that observed for Puf4p (35), which also binds to a 9-nt RNA sequence (Fig. 4B).
Fig. 4.
Defining FBF RNA-binding specificity. (A) Superposition of the CA traces of FBF-2 (red) and PUM1 (yellow) aligning repeats 5–8. (B) Superposition of the CA traces of FBF-2 (red) and yeast Puf4p (green) aligning repeats 5–8. (C) Superposition of the CA traces of FBF-2 (red) and N-terminal (blue) and C-terminal (yellow) halves of PUM1. The separated halves of PUM1 were individually aligned with the corresponding region of FBF-2. The 45-residue region of FBF-2 that transfers 9-nt specificity to PUF-8 is shown in green and contains the hinge point between repeats 4 and 5. DynDom analysis also suggests that there are 2 smaller angle changes between repeats 3 and 4 and repeats 5 and 6. In C, the FBF-2 structure is rotated 15° about the y axis relative to A and B.
The flatter curvature is attributable to the central region of the protein, including repeats 4 and 5. An overall superposition of FBF-2 with PUM1 gives an rmsd of 2.8 Å over 307 CA atoms. The superposition of either the N- or C-terminal half of the proteins separately—either repeats 1–4 or repeats 5–8—yields an rmsd of 1.4 Å (156 N-terminal CA atoms or 161 C-terminal CA atoms) (Fig. 4C). The angle between N- and C-terminal halves in FBF-2 is increased by ≈20° relative to PUM1. We also quantified curvature by measuring the angles between successive equivalent helices along the proteins' long axes (Fig. S6). [As landmarks, we used the CA atoms of residues that would typically form stacking interactions (Fig. S6A).] The repeat-to-repeat angles of FBF-2 are larger than those of PUM1, consistent with its being a flatter RNA-binding surface. Importantly, the major change in FBF-2's curvature is focused between repeats 4 and 5, confirming that this area is critical (Fig. S6B).
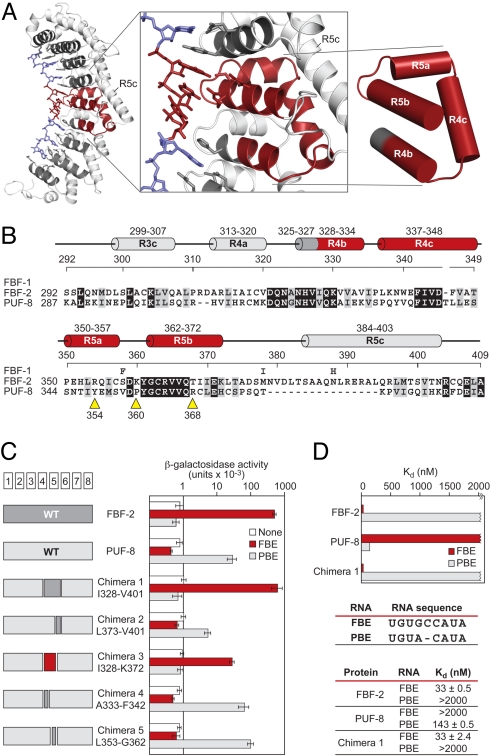
To identify the region of FBF required to impose its specificity, we analyzed chimeras between FBF-2 and PUF-8, a close C. elegans relative of PUM1, using the yeast 3-hybrid system. To monitor RNA binding, we used an FBF-2 site, the gld-1 FBEa, or a PUF-8 site, the PUF-8 binding element (PBE, similar to the NRE). The critical difference between the 2 RNAs is the presence or absence of a single base in the middle of the site (Fig. 5). Near the flipped bases, FBF-2 has 2 distinctive features: an extended third helix in repeat 5 (Fig. 5 A and B, R5c) and a long extended loop between repeats 4 and 5 (Fig. S3, in red). Indeed, a 74-aa fragment of FBF-2 (I328–V401) grafted into PUF-8 generates a chimeric protein with FBF-2's RNA recognition specificity (Fig. 5C, chimera 1) (20). However, a smaller fragment with both the extended helix and loop of FBF-2 does not transfer that specificity, because the chimera still binds the PBE (Fig. 5C, chimera 2) (20). Biochemical assays using purified proteins confirm these findings. Insertion of the 74-aa segment of FBF-2 into a PUF-8 background yielded a protein whose binding to gld-1 FBEa and PBE was essentially indistinguishable from that of FBF-2 (Fig. 5D); similarly, the 74-aa segment abrogated binding of the chimeric protein to the PBE at least 14-fold relative to unmodified PUF-8 (Kd of 143 vs. >2,000 nM; Fig. 5D).
Fig. 5.
Small fragment of FBF opposite the flipped bases imposes its RNA-binding specificity. (A) FBF-2, with the region opposite the flipped bases, is highlighted. Helices in red are the minimal region that imposes the requirement for the extra bases relative to C. elegans PUF-8 (or PUM1). (B) Sequence alignments of FBF-1, FBF-2, and PUF-8 in the minimal region. For FBF-1, we indicate only positions that differ from the FBF-2 sequence. Helices are colored and named as in A. Yellow arrows, sites of single mutations (see the text). Identical residues are shaded in black, and similar residues are shaded in gray. (C) Binding of chimeras. Data (yeast 3-hybrid) are the average of 4 experiments, and error bars are SDs. Chimeras 1–3 used FBF-1, and chimeras 4 and 5 used FBF-2. For simplicity, we use only the FBF-2 numbering. The 2 proteins are identical in the minimal region except for residue 358. (D) In vitro binding analyses of FBF-2, PUF-8, and chimera 1 to FBE and PBE RNA. GST-fusion proteins of FBF-2, PUF-8, and chimera 1 were analyzed by electrophoretic mobility shift assay.
The minimal region of FBF-2 that transferred specificity was a 45-aa fragment (I328–K372) containing portions of PUF repeats 4 and 5 outside the extended helix and loop (red in Fig. 5, chimera 3). Subsections of this region did not alter specificity but, instead, bound the PBE (Fig. 5C, chimeras 4 and 5). The packing of α-helices in the 45-aa region likely yields a flatter curvature; however, swapping single amino acid identities between PUF-8 and FBF-2 did not alter either protein's RNA specificity (Fig. 5B, yellow triangles).
The minimal segment of FBF-2 (I328–K372) lies directly opposite the flipped bases (Fig. 5A, red). We conclude that this fragment of FBF-2 is sufficient to alter the structure of PUF-8 to allow it to bind tightly to an FBF-2 target sequence, imposing the requirement for the extra base. We propose that this change in specificity reflects the imposition of flattened curvature and base flipping.
Discussion
FBF is one of the best-characterized PUF family proteins. Multiple endogenous mRNA targets have been identified and validated, and the biological effects of regulation have been determined directly. Our data reveal how FBF binds these targets.
FBF recognizes 4 natural target sequences similarly. Two sets of bracketing interactions surround a central region in which bases flip away from the protein. The bracketing 5′-U1-G2-U3 and 3′-A7-U8 elements are recognized identically in different RNAs. In contrast, bases 4–6, which vary among mRNA targets, stack with each other and flip away from the RNA-binding surface. Position 9 is poorly conserved and interacts differently depending on the base.
The FBF-2 RNA-binding surface is extended relative to PUM1 because of FBF-2's decreased curvature. The structure of the RNA bound to the protein is adapted to this flattened RNA-binding surface; flipping of bases arises to accommodate the RNA to that surface. An RNA backbone spacer is minimally required to present conserved 5′ and 3′ elements to the elongated surface; even an abasic site at the fifth position binds well (20). This model assumes that the curvature of FBF is the same in the absence of RNA. Although we have been unsuccessful at obtaining crystals of apoFBF-2, comparisons of crystal structures of PUM1 and Puf4p with and without RNA show no change in protein structure in the 2 forms (18, 19, 35, 36).
Flipped bases may be recognized by proteins that bind FBF, such as NANOS-3, GLD-3, and CPB-1, each of which binds RNA as well (5, 13, 15, 37–39). For example, the presence of 3 consecutive flipped bases may enable protein partners to differentiate FBF-containing complexes from those formed with other PUF proteins; indeed, the identity of flipped bases could even allow discrimination of specific subsets of mRNAs seen by FBF. The solvent-exposed hydrophobic face of the sixth base provides an attractive site for interaction. Flipped bases could order assembly of higher order complexes with PUF/RNA complexes at their core. Precedent exists for the role of flipped bases in such assemblies. For example, in the λ-phage transcription antitermination complex, N protein binds to nut site RNA, which is structurally identical to a GAAA tetraloop but with an extruded guanine between the third and fourth positions (40). The extruded base is not required for N protein binding but is essential for subsequent binding of NusA and assembly of an antitermination complex.
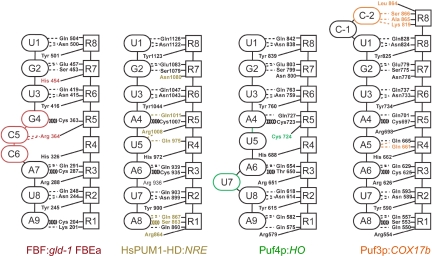
The structures of 4 different PUF protein complexes with RNA (refs. 19 and 35, this work, and companion article in this issue of PNAS) resolve an enigmatic problem; namely, how do different PUF proteins sharing the same amino acids that contact RNA achieve specificity? The atomic interactions of repeats 6–8 with a 5′-UGU and of repeats 2–3 with a downstream AU dinucleotide are essentially the same in every case (Fig. 6). Base flipping is superimposed on that pattern to yield specificity. Indeed, of the 20 atomic contacts between side chains and RNA in FBF, only 3 are different in FBF than in either PUM1 or yeast Puf4p. Yeast Puf4p bound to target RNA and PUM1 bound to noncognate sites also flip bases away from the protein (35, 36), although at different locations within the sites. Thus, the position of flipped bases with respect to the conserved 5′-UGU and 3′-AU sequences adds another layer of specificity. Because the identity of flipped bases is not critical for binding, flexibility in sequence preference at a specific internal position may be a prima facie indicator of a flipped base.
Fig. 6.
Conservation and adaptation of PUF protein–RNA interactions. Schematic representations of interactions between PUF proteins and their RNA targets. Interactions and RNA base conformations unique to each protein are indicated by color: FBF-2 (red), PUM1 (gold), Puf4p (green), and Puf3p (orange).
We suggest that the change in curvature of the proteins creates a requirement for a longer binding site; bases flip to accommodate simultaneous occupancy of the 5′-UGU and 3′-AU binding pockets in repeats 6–8 and 2–3. PUM1 provides the simplest arrangement, in which every base is recognized by cognate amino acids in a helix opposite and no bases are flipped. For FBF and yeast Puf4p, changes in curvature extend the RNA-binding surface. For FBF, the change in curvature is centered between repeats 4 and 5, whereas for Puf4p, it lies between repeats 3 and 4 (35). The structural perturbations impose sequence specificity, as predicted from molecular genetics (20). We propose that PUF proteins likely exist with greater flexibility in the key region, which would then be able to accommodate different modes of binding. Although PUM1 sites possess 8 bases, a ninth base can be accommodated in noncognate sites via base flipping (36). This may represent the nature of intermediates in the evolution of natural PUF specificities.
Although base flipping is critical, it is not the only natural solution to achieving different PUF specificities. In yeast Puf3p, a binding pocket for a cytosine-positioned 2 nucleotides 5′ of the conserved 5′-UGU sequence results in a strict requirement for this base for in vitro binding and in vivo gene regulation (see companion article in this issue of PNAS). The unique specificities of Puf3p, which requires the upstream cytosine, and Puf4p, which requires an extra base, allow multiple PUF proteins to regulate their specific targets in the same organism.
Like yeast, C. elegans expresses multiple PUF proteins, each with unique specificity (20, 28, 41, 42). In addition to FBF and PUF-8, the specificities of PUF-5/6 and PUF-11 have been explored recently (41, 42). PUF-5 recognizes a consensus sequence (5′-CyCUGUAyyyUGU-3′, where y indicates pyrimidine) that includes conserved elements upstream of the 5′-UGU and a downstream UGU rather than AUA (42). The specificity of PUF-11 is particularly intriguing because it binds to 3 different classes of RNA targets, requiring at least 2 different modes of binding (41). One class contains 8 bases (5′-CUGUGAAUA-3′), and 2 classes contain 9-base targets (5′-CUGUANAAUA-3′ and 5′-nUGUnAAAUA-3′) with extra bases inserted at different positions relative to conserved elements. Two of these sequences also appear to require an upstream cytosine at the −1 position. Additional studies are needed to clarify how C. elegans PUF proteins bind their respective target sequences and to identify the full range of RNA targets in vivo.
Natural binding sites for other PUF proteins can be reexamined in light of the principles revealed here. For example, yeast Mpt5p binds 5′-UGUAA-U/C-A-U/A-UA-3′, as inferred from the presence of this sequence in many coimmunopurified RNAs (29). Although neither biochemical nor structural analysis has been performed, we now suggest that the sixth (U/C) and eighth (U/A) bases are excluded from the RNA-binding surface of Mpt5p. We anticipate combinations of flipped bases at other locations in the PUF family, greatly diversifying their specificities. PUF proteins already have been designed with altered specificities, based on the conserved edge-on interactions with bases (20–22). We suggest that it will be possible to design and select proteins that impose requirements for additional flipped bases. Indeed, our chimeras (Fig. 5) define a 45-aa module that can impose the requirement for an additional base.
The requirement for flipped nucleotides has important biological consequences. It enables PUF proteins to discriminate different sets of RNAs yet use an almost identical constellation of atomic contacts (Fig. 6), and it may enable ordered assembly of higher order complexes. Unique specificities can arise during natural or artificial selection simply through a change of curvature rather than through changes in the amino acid residues that contact the RNA. This provides rich opportunities for the evolution of new specificities and networks of control.
Methods
Detailed methods for all procedures are available in SI Text.
Protein Expression and Purification.
The RNA-binding domain of FBF-2 (amino acids 164–575) was expressed as a GST fusion, released by TEV protease cleavage, and purified with a Hi-Trap Heparin column (GE Healthcare). FBF-2/RNA complexes were purified with a Superdex 200 gel filtration column (GE Healthcare).
Crystallization, Structure Determination, and Refinement.
Crystals of FBF-2/RNA complexes were grown by hanging drop vapor diffusion at room temperature with either crystallization solution 1 [100 mM Bicine (pH 9.0), 2% (vol/vol) 1,4-Dioxane, 10% (wt/vol) polyethylene glycol 20,000, 10 mM MgCl2] or solution 2 [100 mM Tris (pH 7.5), 10% (wt/vol) polyethylene glycol 8,000, 8% (vol/vol) ethylene glycol]. Crystals were cryoprotected with 20% (vol/vol) glycerol.
Diffraction data were collected from crystals at 100 K at SER-CAT beamline 22-ID or 22-BM at the Advanced Photon Source, Argonne National Laboratory. The structure of the FBF-2/FBE complex was phased using the SAD method. Crystal structures of the other FBF-2/RNA complexes were determined by molecular replacement using the FBF-2 coordinates from the FBF-2/FBE complex as the search model. Data collection and refinement statistics are shown in Table S1.
Electrophoretic Mobility Shift Assays.
Equilibrium dissociation constants of FBF-2/RNA complexes were determined by electrophoretic mobility shift assay. Apparent dissociation constants were calculated assuming a Hill coefficient of 1. The dissociation constants and SEs reported here were from at least 3 independent experiments. A representative experiment is shown in Fig. S7. The conditions for the assay were established by testing the effect on Kd of varying incubation time (2–48 h), RNA concentration (30–1,000 pM), and time after addition of loading buffer (5–25 min). No significant difference in Kd was detected under these conditions (P ≥ 0.5). Determination of the percentage of active protein was performed as described elsewhere (21). FBF-2 was 85–98% active, GST–FBF-2 was 45–77% active, GST–PUF-8 was 49–70% active, and GST-chimera 1 was 66–77% active. Kd values in Table 1 and Fig. 5D were adjusted based on % active protein.
Yeast 3-hybrid assays.
RNA–protein interactions were analyzed as described by Hook et al. (43).
Supplementary Material
Acknowledgments.
We are grateful to J. Williams and the National Institute of Environmental Health Sciences Protein Microcharacterization Facility for mass spectrometry analyses; J. Gonczy, L. Pedersen, and J. Krahn for crystallographic and data collection support; G. Kissling and Y. Lu for statistical support; and L. Vanderploeg and A. Steinberg for graphics. We are grateful to C. Stumpf for discussions and help with figures. We appreciate our colleagues' comments on the manuscript. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (to T.H.) and by extramural grants from the National Institutes of Health (to M.W.). The Advanced Photon Source used for this study was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31–109-Eng-38.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3K5Q, 3K5Y, 3K5Z, 3K61, 3K62 and 3K64).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812076106/DCSupplemental.
References
- 1.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 2.Wharton RP, Aggarwal AK. mRNA regulation by Puf domain proteins. Sci STKE. 2006;2006:pe37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- 3.Crittenden SL, Kimble J. Analysis of the C. elegans germline stem cell region. Methods Mol Biol. 2008;450:27–44. doi: 10.1007/978-1-60327-214-8_2. [DOI] [PubMed] [Google Scholar]
- 4.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 6.Kaye JA, Rose NC, Goldsworthy B, Goga A, L'Etoile ND. A 3′UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61:57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suh N, et al. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 9.Macdonald PM. The Drosophila pumilio gene: An unusually long transcription unit and an unusual protein. Development. 1992;114:221–232. doi: 10.1242/dev.114.1.221. [DOI] [PubMed] [Google Scholar]
- 10.Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 11.Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992;6:2312–2326. doi: 10.1101/gad.6.12a.2312. [DOI] [PubMed] [Google Scholar]
- 12.Jackson JS, Jr, Houshmandi SS, Lopez Leban F, Olivas WM. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA. 2004;10:1625–1636. doi: 10.1261/rna.7270204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraemer B, et al. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- 14.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14:2596–2609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 17.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 20.Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- 21.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci USA. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- 23.Crittenden SL, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 24.Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Lee MH, Hook B, Lamont LB, Wickens M, Kimble J. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. EMBO J. 2006;25:88–96. doi: 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MH, et al. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson BE, et al. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development. 2005;132:3471–3481. doi: 10.1242/dev.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris AR, Mukherjee N, Keene JD. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White EK, Moore-Jarrett T, Ruley HE. PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA. 2001;7:1855–1866. [PMC free article] [PubMed] [Google Scholar]
- 32.Nakahata S, et al. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J Biol Chem. 2001;276:20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- 33.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson JS, et al. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller MT, Higgin JJ, Hall TM. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol. 2008;15:397–402. doi: 10.1038/nsmb.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta YK, Nair DT, Wharton RP, Aggarwal AK. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008;16:549–557. doi: 10.1016/j.str.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Crittenden SL, et al. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos Trans R Soc London B. 2003;358:1359–1362. doi: 10.1098/rstb.2003.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckmann CR, Crittenden SL, Suh N, Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckmann CR, Kraemer B, Wickens M, Kimble J. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell. 2002;3:697–710. doi: 10.1016/s1534-5807(02)00322-2. [DOI] [PubMed] [Google Scholar]
- 40.Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 41.Koh YY, et al. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hook B, Bernstein D, Zhang B, Wickens M. RNA-protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA. 2005;11:227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeLano W. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.