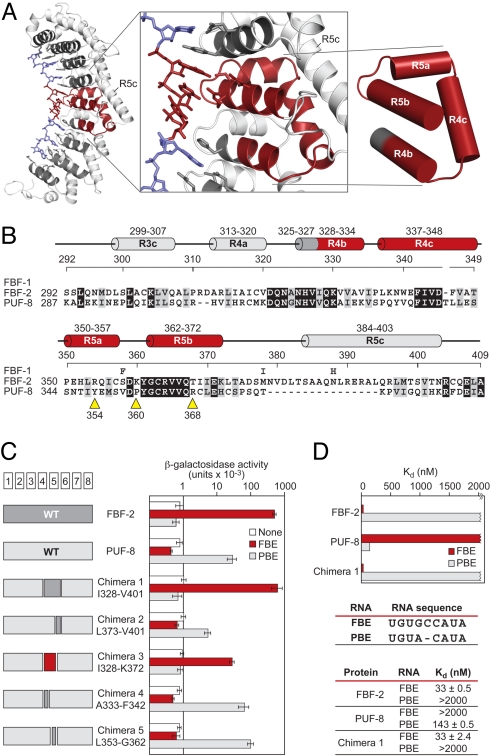
Fig. 5.
Small fragment of FBF opposite the flipped bases imposes its RNA-binding specificity. (A) FBF-2, with the region opposite the flipped bases, is highlighted. Helices in red are the minimal region that imposes the requirement for the extra bases relative to C. elegans PUF-8 (or PUM1). (B) Sequence alignments of FBF-1, FBF-2, and PUF-8 in the minimal region. For FBF-1, we indicate only positions that differ from the FBF-2 sequence. Helices are colored and named as in A. Yellow arrows, sites of single mutations (see the text). Identical residues are shaded in black, and similar residues are shaded in gray. (C) Binding of chimeras. Data (yeast 3-hybrid) are the average of 4 experiments, and error bars are SDs. Chimeras 1–3 used FBF-1, and chimeras 4 and 5 used FBF-2. For simplicity, we use only the FBF-2 numbering. The 2 proteins are identical in the minimal region except for residue 358. (D) In vitro binding analyses of FBF-2, PUF-8, and chimera 1 to FBE and PBE RNA. GST-fusion proteins of FBF-2, PUF-8, and chimera 1 were analyzed by electrophoretic mobility shift assay.