Abstract
The human toll-like receptor 4 (TLR4) pathway is activated in response to lipopolysaccharide (LPS), and subsequent signal transductions lead to the production of cytokines such as tumor necrosis factor-α (TNF-α) by innate immune cells. Defects in innate immune response may contribute to the overproduction of TNF-α leading to systemic inflammation and diseases. Thus, the innate immune response needs to be tightly regulated by elaborate mechanisms to control its onset and termination. LPS tolerance is a state of hyporesponsiveness to subsequent LPS challenge and is achieved by monocytic cells after prolonged exposure to LPS. In this report, kinetics of endotoxin-responsive microRNAs expression analysis revealed a unique pattern of gradual increase for miR-146a starting 4 h after LPS stimulation in THP-1 cells and continued up to 35-fold over 24 h. Conversely, TNF-α increased up to 4 h and then decreased gradually implicating a negative correlation with miR-146a progression. The characteristic up-regulation of miR-146a toward subsequent LPS challenge in THP-1 cells was studied. Strikingly, microRNA expression analysis during the tolerized state of THP-1 cells showed only miR-146a overexpression suggesting its important role in LPS tolerance. In addition, LPS tolerance was dependent on a LPS-priming dose and associated miR-146a up-regulation. LPS-tolerized cells were observed to regain responsiveness in TNF-α production 22 h after LPS removal correlating with a decrease in miR-146a level. Transfection of miR-146a into THP-1 cells mimicked LPS priming, whereas transfection of miR-146a inhibitor largely abolished LPS tolerance. Thus our studies demonstrated that miR-146a is critical for the in vitro monocytic cell-based endotoxin tolerance.
Introduction
Innate immunity plays an important role in providing primary defense against invading pathogenic microorganisms by identifying their conserved components known as pathogen-associated molecular patterns. During infection, pathogen-associated molecular patterns are recognized by the host through several conserved pattern recognition receptors presented on innate immune cells such as monocytes/macrophages and dendritic cells. Toll-like receptors (TLRs)4 are the best characterized and evolutionary conserved pattern recognition receptors, and they play a central role in the initiation of innate immune response by binding to their respective ligands. All TLRs have conserved Toll and IL-1 receptor domain in the cytosolic region, which activates common signaling pathways, most notably through activation of NF-κB transcription factor.
Lipopolysaccharide (LPS or endotoxin) is the principal component of the outer membrane of Gram-negative bacteria. LPS-induced TLR4 signal transduction activates NF-κB, leading to the production of pro-inflammatory cytokines such as IL-1β and TNF-α (1). Pathological dysregulation of NF-κB is linked to inflammatory diseases such as sepsis, autoimmune diseases, and possibly cancer (2). Neutrophils and monocytes from sepsis patient are refractory to subsequent LPS challenge and no longer produce these cytokines (3). This phenomenon, referred to as endotoxin tolerance, is also a mechanism to prevent overstimulation from the continuous exposure to same danger signals in the environment. Endotoxin tolerance has been established for decades in vivo (4) and has also been extensively investigated in vitro using primary monocytes/macrophage cells and cell lines (5–8). To understand the endotoxin tolerance mechanism, changes of cell surface molecules, signaling proteins, pro-inflammatory and anti-inflammatory cytokines, and other mediators have been studied. Despite intense investigations for decades into the hyporesponsiveness associated with innate immune cells in response to LPS priming, there is no consensus yet on the primary mechanism responsible for its development (1).
MicroRNA (miRNA) is a new class of regulators of gene expression that acts at the post transcriptional level via an RNA interference mechanism (9). In mammals, miRNA biogenesis involves the initial transcription by RNA polymerase II of primary miRNAs, which are sequentially cut by two RNase III enzymes, Drosha and Dicer, and create ∼23-nucleotide double-stranded RNA duplexes (10). Eventually, the mature miRNA guide strand is loaded into the miRNA-induced silencing complex, where it guides the recognition and translational repression or degradation of target mRNAs. miRNAs have emerged to play important roles in many biological processes ranging from cellular development and differentiation to tumors (9). Recently, miRNAs have been shown to be involved in innate immunity. During the activation of an innate immune response, a rapid increase in the expression of selected miRNAs, namely miR-146a, miR-132, and miR-155 (11), miR-125a (12), and miR-9 (13) have been observed in monocytic cell lines or mouse macrophages in response to LPS, but their biological activities are still obscure, and many studies are needed on their kinetics and subsequent putative role in innate immunity. Initial studies on miR-146a expression in response to microbial components and cytokines, including IL-1β indicate that it is involved in innate immunity against bacterial pathogens and is also implicated in inflammatory diseases. Interestingly, this response does not seem to be restricted to inflammatory cells, because miR-146a expression has been observed in lung epithelial cell (14). Further analysis, to determine the biological role of miRNA-146a, reveals that its expression is NF-κB-dependent and regulates production of cytokines such as IL-1β and TNF-α in innate immunity (11). IL-1 receptor-associated kinase (IRAK-1) and TNF receptor-associated factor-6 (TRAF6), which are important in TLR4 and pro-inflammatory cytokine (IL-1) signaling, have been established as molecular targets for miR-146a (11). More importantly, IRAK-1 and TRAF6 are known to be part of the common signaling pathway derived from TLR-2, -4, and -5 and the IL-1β receptor, leading to speculation that increased miR-146a expression might act in a negative feedback pathway. Previously, Li et al. (15) and Boone et al. (16) observed LPS tolerance in monocytes caused by impairment of IRAK-1 and TRAF6 kinase activity, respectively. Considering its ability to regulate TRAF6 and IRAK-1, we hypothesize that miR-146a to be involved in endotoxin tolerance. However, association between miR-146a overexpression and endotoxin tolerance has not been examined.
The aim of this study is to investigate the unique expression pattern of miR-146a compared with other LPS-responsive miRNAs and to observe its role in tolerance in innate immune cells to subsequent LPS exposure. Our findings suggest that miR-146a contributes to controlling TNF-α production and provides innate immunity to evade recurrent bacterial infection through establishing endotoxin tolerance.
EXPERIMENTAL PROCEDURES
Cell Culture and LPS Stimulation
Human THP-1 cells, an undifferentiated promonocytic cell line, were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained by twice weekly passage in RPMI 1640 medium containing 25 mm HEPES and l-glutamine (BioWhittaker, Lonza, Walkersville, MD), 10% fetal bovine serum (Mediatech Inc., Manassas, VA), and 100 units/ml penicillin-streptomycin (Mediatech) at 37 °C with 5% CO2. Log phase cells were used in all experiments and cultured at a density of 106 cells/ml.
To determine the kinetics of LPS-induced cytokine production in vitro, fresh THP-1 cells were suspended in complete RPMI 1640 culture medium and seeded at 106 cells/ml in a 24-well plate. Cells were stimulated with 0, 10, 100, and 1000 ng/ml highly purified Salmonella enterica serotype Minnesota LPS (Sigma) prepared in the same culture medium by serial dilution of a 500 μg/ml stock solution in tissue culture grade phosphate-buffered saline (PBS). Cells were harvested and supernatants were collected at 2, 4, 8, 12, or 24 h after incubation and stored at −80 °C until assayed for cytokine levels. Cell pellets were washed in PBS and stored in RNAlater at 4 °C for total RNA isolation in subsequent analysis.
In Vitro Induction of Endotoxin Tolerance
An LPS tolerance cell model using monocytic cell line THP-1 was adapted from methods described previously with some minor modifications (17–19). Briefly, before starting tolerance assay, THP-1 cells were cultured for ∼4 days until cells were in log phase and concentration at 106 cells/ml and viability was checked by trypan blue staining to be >99%. THP-1 cells (5 × 105 cells/ml) were transferred in fresh complete medium in new 5- or 25-ml flasks. The cells were incubated with low dose of LPS (10 ng/ml) for 18 h. In some studies, cells were primed with 10, 100, or 1000 ng/ml LPS. After two washes with tissue culture grade PBS, cells were resuspended in complete culture medium alone or with LPS (0, 10, 100, or 1000 ng/ml, final concentration) and distributed in 24-wells plate and returned to the CO2 incubator for an additional 5 h at 37 °C. Supernatants were harvested and stored at −80 °C until assayed for TNF-α.
Quantification of miRNA and mRNA Expression Level by qRT-PCR
Total RNA of THP-1 cells was isolated using the mirVana isolation kit (Ambion, Austin, TX) following the manufacturer's protocol. RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technology Inc., Wilmington, DE), and equal amounts of each RNA (6.7 ng for miRNA and 33 ng for mRNA) were used for real-time RT-PCR (qRT-PCR) analysis. miRNAs analysis were performed using the TaqMan MicroRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix, and TaqMan microRNA Assay primers for human miRNAs (Applied Biosystems, Foster City, CA). For mRNA analysis, a High Capacity cDNA RT Kit (Applied Biosystems) and TaqMan mRNA assay primers for IRAK-1 and TRAF6 were used. The cycle threshold (Ct) values, corresponding to the PCR cycle number at which fluorescence emission reaches a threshold above baseline emission, were determined, and miRNA expression values were calculated using RNU44 as endogenous control (Applied Biosystems) following the 2−ΔΔCt method (20). mRNA expression values were quantified in the same way after normalization to 18 S RNA.
THP-1 Cell Transfection
Pre-miR miRNA precursor molecules (miR-146a mimic) and anti-miR miRNA inhibitors (miR-146a inhibitor) were purchased from Ambion and dissolved in nuclease-free water, and the resulting 20 μm stock was stored in aliquots at −80 °C prior to use. One day before the transfection, cells were transferred in fresh culture medium at a concentration of 5 × 105 cells/ml. The following day, THP-1 cells (5 × 105 cells/well) were transfected with 40 nm of precursor or inhibitor using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. miR-146a mimic-transfected THP-1 cells were incubated for 24 h followed by washing two times with complete growth medium. The washed cells were treated with 1000 ng/ml LPS for 3 h. For miR-146a inhibitor experiment, transfected THP-1 cells were incubated for 24 h followed by washing with complete growth medium and addition of 10 ng/ml LPS for an additional 16 h. Cells were then washed with complete growth medium and challenged with 1000 ng/ml LPS for 3 h. Supernatants from cell cultures were collected and assayed for TNF-α secretion, and cell pellets were used for RNA isolation and real-time PCR analysis.
ELISA for TNF-α
Supernatants were collected from cell cultures at different time points after stimulation with various concentrations of LPS. Secreted TNF-α protein in the supernatants was measured by enzyme-linked immunosorbent assay (ELISA) using an Opt EIA TNF-α kit as recommended by the manufacturer (BD Biosciences). Absorbance was measured at 405 nm using a microplate reader (Model 680, Bio-Rad). A405 was converted into concentrations using standard curves of recombinant human TNF-α.
IL-1β and IL-6 Assays
Cultured supernatants were analyzed using the human cytokine/chemokine Milliplex MAP kit (Millipore) according to the manufacturer's protocol to quantitatively detect IL-1β and IL-6. Samples were analyzed on a 200 system (Luminex, Austin, TX).
Western Blot Analysis
LPS-tolerized and untolerized THP-1 cells (5 × 106/condition) were collected 2 h after LPS challenge, pelleted at 1000 × g for 10 min and lysed on ice for 10 min in 1 ml of lysis buffer (50 mm HEPES, pH 7.6, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 20 mm β-glycerophosphate, 1 mm sodium orthovanadate, 1 mm sodium fluoride, 1 mm benzamidine, 5 mm para-nitrophenyl phosphate, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and Complete protease inhibitor mixture from Roche Diagnostic, Indianapolis, IN). Soluble lysates were quantitated for protein concentration using a Bio-Rad protein assay kit, separated by SDS-PAGE (10% acrylamide) along with a broad range molecular weight markers (Bio-Rad), and electroblotted to a polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked overnight at 4 °C with 5% nonfat milk in 1× PBS/0.05% Tween 20 (PBS-T) and were probed with primary rabbit anti-IRAK-1 or anti-TRAF6 at 1:200 (Santa Cruz Biotechnology, Santa Cruz, CA). The membranes were washed three times with PBS-T and incubated for 1 h with goat anti-rabbit IgG-horseradish peroxidase at 1:5000 (Southern Biotechnology, Birmingham, AL). After washing in PBS-T, reactive bands were visualized by Super Signal chemiluminescent reagent (Pierce).
Statistical Analysis
Data are presented in the figure as mean ± S.E. For multiple group comparisons, one-way analysis of variance (p < 0.05) was performed, followed by the two-sided, unpaired Student's t test as described by Shaffer (21). Unpaired two-tailed Student's t test was used to compare two independent groups. For all statistical analysis, Prism for Windows, version 5.0 (GraphPad Software, San Diego, CA) was used, and p < 0.05 was considered statistically significant.
RESULTS
Kinetics of LPS-induced Expression of TNF-α, miRNAs, and Adaptor Kinases
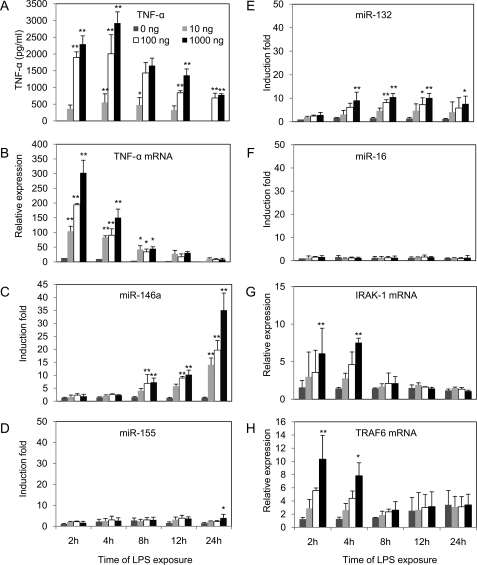
LPS is one of the potent stimulator of monocytes and macrophages in innate immunity, and it induces the production of a diverse array of inflammatory mediators, including TNF-α both in vitro and in vivo (22). To observe TNF-α production in vitro, the most commonly employed monocytic cell line, THP-1 (23), was used in the present study. After LPS stimulation of THP-1 cells, kinetics of TNF-α production was examined. As shown in Fig. 1A, log phase THP-1 cells were treated with LPS at concentrations of 0, 10, 100, or 1000 ng/ml and harvested at different time points from 2 to 24 h to assess TNF-α production as measured by ELISA described under “Experimental Procedures.” In supernatants from cultured LPS-stimulated monocytes, TNF-α started to appear within 2 h and reached a maximal level at 4 h of stimulation followed by gradual decrease starting at 8 h. Highest dose of LPS (1000 ng/ml) generated the highest level of TNF-α up to 3000 pg/ml at 4 h, consistent with levels reported previously (17). The progressive changes in TNF-α level was validated at the mRNA level by qRT-PCR analysis performed on RNA samples collected from the same cells shown in Fig. 1B. The kinetics of TNF-α mRNA expression showed similar progression compared with secreted TNF-α protein except the peak level of TNF-α mRNA was 2 h (versus 4 h peak of protein).
FIGURE 1.
Dose- and time-dependent expression of TNF-α and adaptors TRAF6 and IRAK1 mRNA preceded that of miR-146a in LPS-stimulated monocytic THP-1 cells. THP-1 cells were treated with 0, 10, 100, or 1000 ng/ml LPS and incubated for 2, 4, 8, 12, or 24 h. Culture supernatants were collected at the indicated time points from 2 to 24 h for TNF-α protein analysis using ELISA (A). Total RNA were purified from the respective cell pellets and analyzed by qRT-PCR for the expression of TNF-α mRNA (B), miR-146a (C), miR-155 (D), miR-132 (E), miR-16 (F), IRAK-1 mRNA (G), and TRAF6 mRNA (H). mRNA and miRNA expression were normalized with control 18 S RNA and RNU44, respectively. All results are expressed as mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01 compared with untreated cells.
LPS causes up-regulation of miR-146a in THP-1 cells and monocytes as demonstrated by Taganov et al. (11) and Bazzoni et al. (13) independently, through microarray analysis. Following their observations and considering the subsequent experimental purposes in this study, kinetics of miR-146a, miR-155, and miR-132 expression were determined by qRT-PCR analysis on the same RNA samples (Fig. 1, C–F). The -fold changes in miRNA expression were calculated by comparing the value of LPS-treated cells to that of untreated samples cultured in parallel. miR-146a showed an average increase of 7-fold after 8 h, and interestingly a gradual increase was observed up to 35-fold over 24 h (Fig. 1C). In contrast, miR-155 and miR-132 expression showed increases of 4- and 10-fold, respectively, after 12 h of LPS treatment, and no further increase was observed in 24 h (Fig. 1, D and E). No significant change in the expression of miR-16 was observed (Fig. 1F), and this could be considered as an additional LPS-independent control together with the internal control RNU44. Most strikingly, in response to LPS, miR-146a is the only LPS-induced miRNA that showed a gradual increase significantly over 24 h (Fig. 1C). In one set of experiments, miR-146a continued to elevate up to 45-fold at 48 h after LPS treatment (data not shown).
Target prediction algorithms have shown that two important adaptor molecules IRAK-1 and TRAF6 are two of the top targets for miR-146a. This was experimentally validated by Taganov et al. (11) by demonstrating miR-146a-induced reduction of luciferase expression in luciferase reporters containing 3′-untranslated repeat of these adapter molecules. Fig. 1, G and H, show the respective increases in IRAK-1 and TRAF6 at 2 and 4 h and decreases at 8 h and subsequent time points in line with the elevated expression of miR-146a starting at 8 h. These data are consistent with IRAK-1 and TRAF6 being regulated by miR-146a in NF-κB-driven inflammatory response to LPS.
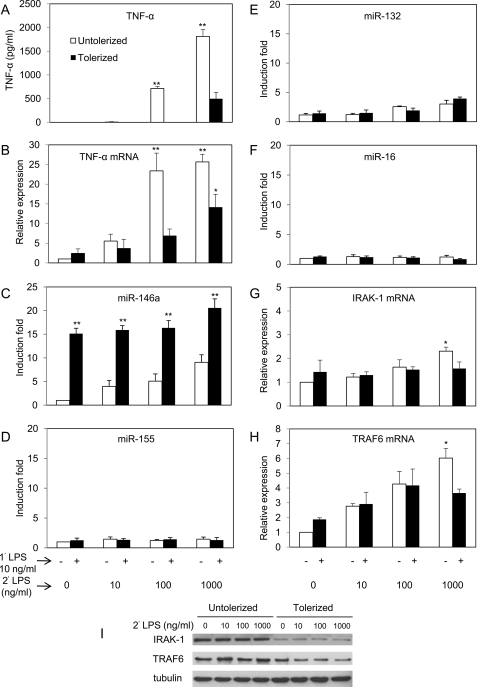
Pretreatment of THP-1 Cells with LPS Results in Reduced TNF-α Production Linked to miR-146a Overexpression
TNF-α is considered as the most commonly used marker of endotoxin tolerance due to its dramatic reduction following subsequent LPS challenge compared with primary treatment of LPS (24). Fig. 2 shows the ability of LPS to induce tolerance based on TNF-α production using the THP-1 cell model. THP-1 cells were primed with 10 ng/ml LPS for 18 h followed by washing with PBS and challenged with various doses of LPS ranging from 0 to 1000 ng/ml. After 5 h of incubation with challenged LPS, TNF-α protein level was analyzed by ELISA (Fig. 2A). TNF-α production decreased ∼4-fold compared with the untolerized control after challenged with 1000 ng/ml LPS, demonstrating significant tolerance induction as expected from previous studies (17). Analysis of mRNA level of TNF-α by qRT-PCR (Fig. 2B) showed levels consistent with the ELISA data, and reduction of TNF-α mRNA was significant when challenged with either 100 or 1000 ng/ml LPS. Treatment with 10 ng/ml (or 1 ng/ml, data not shown) of LPS in this instance did not show discernable difference in TNF-α protein or mRNA compared with control 5 h after LPS challenge. In one experiment, THP-1 cells were primed with 10 ng/ml TNF-α for 18 h and then challenged with 1000 ng/ml LPS to determine whether TNF-α could substitute LPS priming to induce LPS tolerance. The results showed that TNF-α did not induce tolerance, and it was noted that the level of miR-146a was not significantly above untreated control 18 h after TNF-α treatment (data not shown).
FIGURE 2.
High level of miR-146a may account for LPS tolerance in THP-1 cell model. THP-1 cells primed with 10 ng/ml LPS continuously for 18 h (tolerized, ■) and untreated controls incubated for the same time period (untolerized, □) were washed twice with PBS and challenged with 0, 10, 100, or 1000 ng/ml LPS for 5 h. Culture supernatants and total RNA were analyzed as described under “Experimental Procedures.” A–H, all results are expressed as mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01 compared with untolerized THP-1 cells. Lysates of the tolerized and untolerized cells 2 h after LPS challenge were analyzed for IRAK-1, TRAF6, and tubulin expression by Western blot (I).
To understand the role of miRNA in LPS tolerance, miRNA analyses in the tolerized THP-1 cells were performed in relation to the untolerized control shown in Fig. 2 (C–F). As expected, miR-146a in the tolerized sample showed significantly higher expression over 18 h of initial incubation plus 5 h of challenge, whereas little or no changes in expression of miR-155, miR-132, and miR-16 were observed compared with untolerized controls. Fig. 2 (G and H) shows the expression of known miR-146a targets IRAK-1 and TRAF6 in both tolerized and untolerized cells. The levels of IRAK-1 and TRAF6 were not affected significantly up to 100 ng/ml of secondary LPS challenge compared with the untolerized controls. At 1000 ng/ml LPS challenge, there was significant reduction in levels of both IRAK-1 and TRAF6. These data suggest that these adaptor molecules might be affected at the post-transcriptional level. Confirmation of the moderate reduction of IRAK-1 and TRAF6 proteins was obtained in the Western blot analysis comparing LPS tolerized to control untolerized cells (Fig. 2I). Consequently, miR-146a may play an important role in LPS tolerance based on the inverse correlation with TNF-α production and levels of IRAK-1 and TRAF6 in tolerized THP-1 cells.
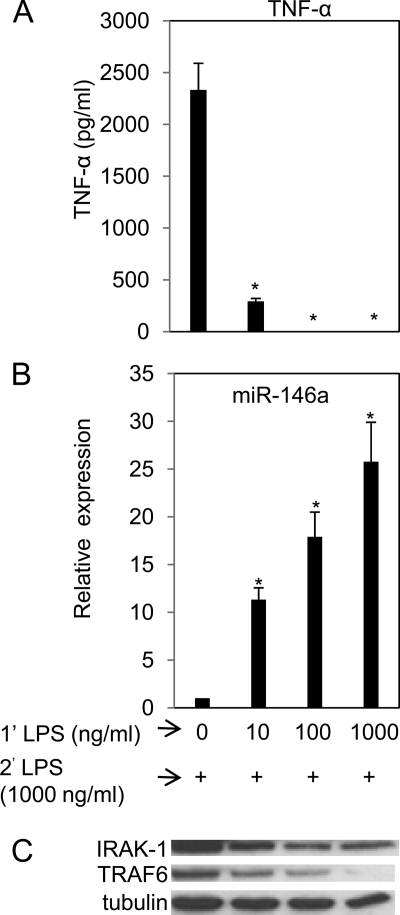
Higher Doses of LPS Priming Render Efficient Tolerance and Higher miR-146a Expression
Endotoxin tolerance is usually studied at a low dose (10 ng/ml) of LPS priming. However, in nature, animals are always exposed to various doses of LPS and are refractory to LPS challenge. To obtain better understanding about the impact of the priming concentration in relation to miR-146a expression on endotoxin tolerance in vitro, a range of priming concentrations from 0, 10, 100, and 1000 ng/ml was employed followed by 1000 ng/ml LPS challenge. In one of the earlier experiments, priming with 1 ng/ml LPS did not show significant difference from control and was not included in subsequent experiments (data not shown). Priming with 10 ng/ml LPS resulted in ∼90% decrease (p < 0.01) of TNF-α production (Fig. 3A). Complete unresponsiveness was detected with 100 and 1000 ng/ml LPS priming. This result is consistent with the previous study by both LaRue et al. (18) and Jacinto et al. (19) where THP-1 cells primed with higher dose of LPS were more refractory to subsequent LPS challenge. Fig. 3B shows gradual augmentation of miR-146a correlated with tolerance but similar changes in other miRNAs such as miR-155 and miR-132 were not observed (data not shown). These data are consistent with Fig. 1C, which showed miR-146a expression to be LPS dose-dependent. Western blot analysis confirmed the moderate, gradual reduction in IRAK-1 and TRAF6 levels with the increase of miR-146a expression proportional to LPS priming dosage (Fig. 3C). Thus, these data showed the positive correlation of LPS priming dose with up-regulation of miR-146a and LPS tolerance.
FIGURE 3.
Higher priming dose of LPS induced higher miR-146a levels and more efficient suppression of TNF-α production in subsequent LPS challenge. THP-1 cells were primed with 0, 10, 100, or 1000 ng/ml LPS continuously for 18 h, washed twice with PBS, and challenged with 1000 ng/ml LPS. Supernatants and cell pellets were collected 5 h later as described under “Experimental Procedures” for TNF-α protein determined by ELISA (A), miR-146a expression analysis in total RNA (B). Data points and error bars represent mean ± S.D. of three independent experiments. *, p < 0.01 compared with untreated cells. Western blot analysis for IRAK-1, TRAF6, and tubulin in the cell lysates collected 2 h after LPS challenge (C).
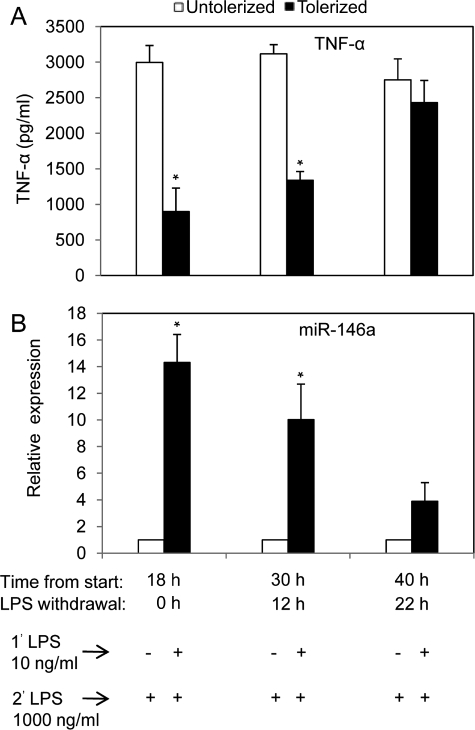
The Elevated Expression of miR-146a Depends on Continued Exposure to LPS and Is Correlated with the Requirement for LPS Tolerance
In the first LPS tolerance experiment (Fig. 2), the results suggested that, among the known LPS-induced miRNA analyzed, the elevated miR-146a expression levels were most correlated with LPS tolerance. When THP-1 cells were continuously exposed to LPS, the level of miR-146a continued to rise at 24 h (Fig. 1C) and 48 h (data not shown). To determine whether the continuing increase in miR-146a level requires continued exposure to LPS, THP-1 cells were primed with 10 ng/ml LPS continuously for 18 h, then washed twice with PBS and cultured in complete growth medium for an additional 0, 12, or 22 h (LPS withdrawal). At each time point, 8 × 105 cells were challenged with 1000 ng/ml LPS for 5 h prior to analysis for TNF-α protein production in culture supernatant by ELISA (Fig. 4A) and qRT-PCR analysis of miR-146a expression (Fig. 4B). With 18 h of continuous LPS priming and 5 h of LPS challenge, the same condition used in Fig. 2A, significant reduction in TNF-α secretion and highly elevated miR-146a level were observed again as expected. Interestingly, after 12 h of LPS withdrawal, cells started to regain LPS responsiveness to TNF-α production and almost completely recovered from tolerance after 22 h of LPS withdrawal (Fig. 4A). Fig. 4B shows the expression of miR-146a decreased 12 and 22 h after LPS withdrawal. At 22 h after LPS withdrawal, the levels of TNF-α and miR-146a were no longer significantly different from the unprimed controls. Thus, the presence of up-regulated miR-146a due to LPS priming is important for maintaining tolerance, because it is diminished gradually with the disappearance of miR-146a in washed THP-1 cells.
FIGURE 4.
Reduction in miR-146a expression in LPS tolerized THP-1 cells inversely correlated with TNF-α production. THP-1 cells were cultured with (tolerized) or without (untolerized) 10 ng/ml LPS continuously for 18 h. Cells were then washed twice with PBS and cultured in complete growth medium for an additional 0, 12, or 22 h (LPS withdrawal). At each time point, 8 × 105 cells were challenged with 1000 ng/ml LPS for 5 h prior to analysis of TNF-α protein production in culture supernatant by ELISA (A) and qRT-PCR analysis of total RNA for miR-146a expression (B). Values are expressed as mean ± S.D. from three independent experiments. *, p < 0.01 compared with LPS untreated cells.
Up-regulation of miR-146a Alone Can Mimic LPS Priming to Induce LPS Tolerance
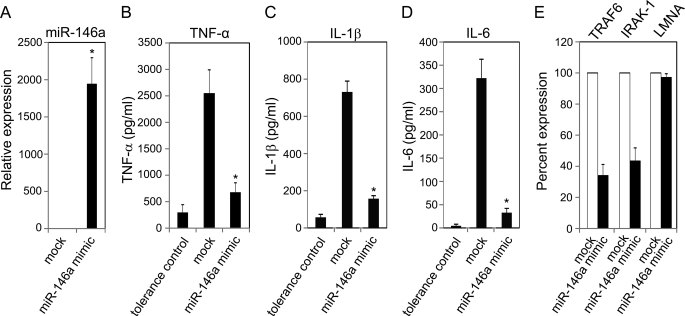
LPS has been shown to induce the expression of a few regulatory miRNAs (11, 13). Similar up-regulation of miR-146a was observed in LPS-primed and LPS-tolerized THP-1 cells in this study. To determine the direct role of miR-146a in endotoxin tolerance, THP-1 cells were transfected with 40 nm miR-146a mimic followed by challenge with 1000 ng/ml LPS after 24 h. miR-146a was expressed 1,850-fold higher in miR-146a mimic transfected cells than the mock transfected cells confirming the successful transfection (Fig. 5A). In Fig. 5B, cells transfected with miR-146a mimic produced ∼4 times less TNF-α in comparison to mock transfected cells 3 h after the LPS challenge. The ability of transfected miR-146a to cause tolerization was almost similar to the tolerance positive control (4- versus 6-fold). Besides TNF-α, other pro-inflammatory cytokines are produced through NF-κB activation and transcriptional regulation. As expected, Fig. 5 (C and D) shows similar changes for two other NF-κB-regulated cytokines IL-1β and IL-6. To determine the effect of overexpression of miR-146a on adaptor molecules, RNA samples from transfected cells were analyzed for the mRNA levels of TRAF6 and IRAK-1, both targets of miR-146a. THP-1 cells transfected with miR-146a mimic showed a 62% reduction of TRAF6 and a 56% reduction of IRAK-1 compared with mock transfected cell (Fig. 5E). An unrelated human gene lamin A/C was also included as an additional control showing no significant changes in expression level between miR-146a mimic transfected and mock control. These data demonstrate the ability of miR-146a alone to induce LPS tolerance via specific negative regulation of TRAF6 and IRAK-1.
FIGURE 5.
Transfected miR-146a alone could mimic LPS priming in LPS tolerance assay. THP-1 cells transfected with 40 nm miR-146a mimic followed by incubation for 24 h, mock transfected cells, and 10 ng/ml LPS primed cells (tolerance positive control) were washed with complete growth medium and challenged with 1000 ng/ml LPS for 3 h. A, RNA isolates were prepared from cell pellets of mock and transfected cells followed by qRT-PCR for miR-146a expression normalized with RNU44. B, ELISA analysis of TNF-α protein in supernatants tolerance-positive control, mock transfected, and miR-146a mimic transfected cells. C and D, multiplex analysis of IL-1β and IL-6 proteins in supernatants of tolerance-positive control, mock transfected, and miR-146a mimic-transfected cells. E, the same RNA samples were also analyzed by qRT-PCR for mRNA expression of miR-146a targets TRAF6, IRAK-1, and an unrelated gene lamin A/C (LMNA, negative control). Data are representative of three independent experiments and expressed as mean ± S.D. *, p < 0.01 compared with mock transfected cell.
Knockdown of miR-146a in THP-1 Cells Diminishes LPS Tolerance
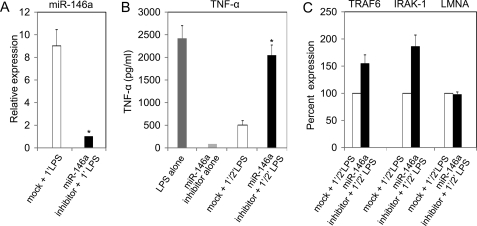
As shown in Fig. 5, transfected miR-146a induced LPS hyporesponsiveness in THP-1 cells. To corroborate the role of miR-146a on LPS tolerance, the effect of miR-146a knockdown on LPS tolerance was determined by suppressing its activity with miR-146a inhibitor. THP-1 cells were transfected with miR-146a inhibitor at a concentration of 40 nm, and supernatant as well as total RNA samples were collected as described under “Experimental Procedures.” Because the level of miR-146a is very low in unstimulated THP-1 cells, the knockdown effect in these cells transfected with the miR-146a inhibitor might not be easily appreciated. Thus to confirm the functional activity of the miR-146a inhibitor, the knockdown of miR-146a was monitored in miR-146a inhibitor-transfected cells to determine whether it would efficiently block miR-146a production upon LPS stimulation. As shown in Fig. 6A, miR-146a expression in transfected cells was down-regulated by 9-fold compared with mock transfected cells primed with LPS. Cells transfected with miR-146a inhibitor showed efficient response to LPS and produced significantly higher (p < 0.01) TNF-α protein compared with mock and mR-146a inhibitor alone (Fig. 6B). THP-1 cells transfected with miR-146a inhibitor showed a 52% increase of TRAF6 and an 80% increase in the level of IRAK-1 compared with mock transfected cells (Fig. 6C). In contrast, the expression of an unrelated gene lamin A/C was not affected in miR-146 inhibitor-transfected cells. Thus, LPS tolerance was disrupted due to knockdown of miR-146a, which helped cells to regain LPS responsiveness. Taken together, these data demonstrate that LPS-induced miR-146a up-regulation is critical to endotoxin tolerance.
FIGURE 6.
Blockade of miRNA-146a expression abrogated LPS-induced tolerance in THP-1 cells. THP-1 cells transfected with 40 nm miR-146a inhibitor for 24 h, along with mock transfected, were washed with complete growth medium and primed with 10 ng/ml LPS for 16 h. Cells were then washed with complete growth medium and challenged with 1000 ng/ml LPS for 3 h. Additional controls included were cells transfected with miR-146a inhibitor alone and cells treated with 1000 ng/ml LPS alone for 3 h (LPS alone, TNF-α-positive control). A, total RNA from cell pellets of mock and miR-146a inhibitor-transfected cells, primed with 10 ng/ml LPS for 16 h, were analyzed by qRT-PCR for miR-146a expression. B, TNF-α protein in supernatants of TNF-α (+) control, miR-146a inhibitor alone (without LPS), mock transfected, and miR-146a inhibitor-transfected, were measured by ELISA. C, total RNA were analyzed for TRAF6, IRAK-1, and an unrelated gene lamin A/C (LMNA) expression in both mock and miR-146a inhibitor-transfected cells by qRT-PCR. Data are expressed as mean ± S.D. of three independent experiments. *, p < 0.01 compared with mock transfected cells.
DISCUSSION
Previously, changes in miRNA expression profiles in THP-1 cells were reported primarily using only a single LPS dose at fixed time, but in this report, kinetics of the LPS-induced miRNA was analyzed using different LPS doses for up to 24 h. A distinct expression pattern was observed for each LPS-induced miRNA, but only miR-146a persisted for an extended period and at very high levels. A similar expression pattern is likely to occur in the host depending on the danger signal. Li et al. (15) and Jacinto et al. (19) reported IRAK-1 protein level dramatically decreased within 90 min after LPS priming of monocytes and remained at low level following 6 h of LPS challenge. Consistent with this finding, a negative correlation between the expression of miR-146a and adaptor kinases IRAK-1 and TRAF6 as well as TNF-α (protein and mRNA) was demonstrated in this study. It is acknowledged that the regulation of IRAK-1 and TRAF6 by miR-146a has been characterized (11) and confirmed in some subsequent studies (25–27). The new data in this report are focused on defining the role of miR-146a in endotoxin tolerance.
Endotoxin tolerance has long been recognized as an interesting mechanism that controls the intensity and duration of innate immune cell activation, but a key regulator for tolerance has not been identified (1). Nomura et al. (28) described stimulation of monocytes with LPS leading to the down-regulation of TLR4-MD2 (myeloid differentiation protein-2) from the cell surface, essentially causing them to be unresponsive during a second LPS challenge. However, in a 2002 review article, Dobrovolskaia and Vogel (29) concluded that much higher doses of LPS were required for the TLR4 receptor down-regulation than for tolerance induction, and sustained overexpression of TLR4 did not prevent tolerance induction (30). In another study, LPS tolerance has been shown to be independent of LPS co-receptor CD14, because THP-1 cells showed tolerance in the presence or absence of anti-CD14 monoclonal antibody (17). Randow et al. (31) reported that anti-inflammatory cytokines such as IL-10 and transforming growth factor-β could lead to deactivation of macrophages in response to LPS. On the other hand, Berg et al. (32) showed that IL-10 and transforming growth factor-β knock-out mice were still capable of developing LPS tolerance. Similarly, LPS tolerance was shown to be independent of other cytokines like interferon-γ because LPS tolerance could be induced in interferon-γ knock-out mice (33). In short, the key regulator for endotoxin tolerance has not been identified, and clearly our finding to report the critical role of miR-146a in this mechanism is novel.
miR-146a Plays a Critical Role in LPS Tolerance
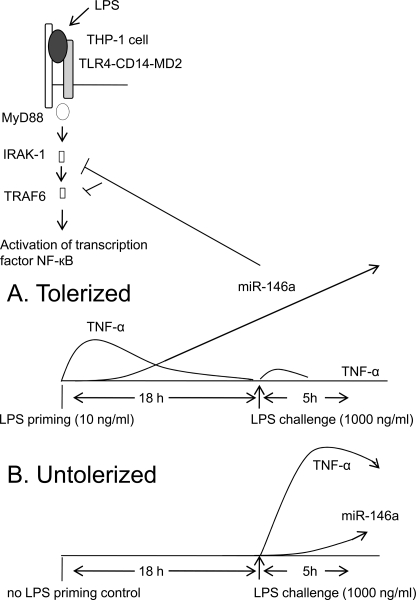
Fig. 7 summarizes our proposed model for the critical role of miR-146a in LPS tolerance in THP-1 cells. LPS binds to the LPS-binding protein, which in turn is coupled to CD14 on the cell surface of monocytes. Subsequently, LPS-CD14 interacts with TLR4 and forms a complex with another accessory protein MD-2. The TLR4 signaling cascade is initiated after binding with adaptor protein MyD88 that acts as a bridge between TLR4 and incoming IRAK-1 adaptor kinase, which further recruits TRAF6. This chain of events triggers activation and translocation of NF-κB and results in the transcription of immune-responsive genes and cytokines such as TNF-α and mRNA regulators such as miR-146a. Consequently, low dose LPS-primed THP-1 cells produce TNF-α rapidly and continue to do so for 4–6 h, and then TNF-α production decreases as soon as regulatory miR-146a starts to increase. Then, 18 h post-priming, a profound difference between miR-146a expression and TNF-α secretion is established, and because the up-regulated miR-146a acts negatively on IRAK-1 and TRAF6 mRNA the cells become tolerized (Fig. 7A). Tolerized cells do not respond to high dose LPS challenge due to the high level of miR-146a unlike the untolerized control, which is responsive to LPS at this stage (Fig. 7B).
FIGURE 7.
A model of the role of miR-146a in LPS-TLR4-mediated signal transduction in tolerized (A) versus untolerized (B) cells. See text under “Discussion.” MD2, myeloid differentiation protein-2.
Priming time is important to render monocytic cells to be tolerant. LaReu et al. (18) have observed moderate tolerance in THP-1 cell as early as 6 h and profound tolerance after 18 h of LPS priming. Consistent with this time frame, even THP-1 cells stimulated with low dose LPS (10 ng/ml) showed high level of miR-146a expression. Furthermore, Jacainto et al. (19), and this report showed that LPS tolerance correlated with the increase in LPS-priming dose and complete tolerance (zero TNF-α production) was observed when 100 ng/ml or higher concentration of LPS was used; these high concentrations of LPS were linked with the high up-regulation of miR-146a confirming its substantial role in LPS tolerance.
The biological significance of other miRNAs induced by LPS such as miR-132 and miR-155 on LPS tolerance is not clear. They might have direct or indirect roles on LPS tolerance together with miR-146a. Thus, transfection studies were performed to examine the effects of overexpression of miR-146a alone on LPS tolerance. Transfection of miR-146a-mimic into THP-1 cells resulted in dramatic reduction of TNF-α indicating its negative regulatory effect on TNF-α production and subsequent LPS challenge. In the transfected cells, miR-146a level was pronounced, although TRAF6 and IRAK-1 mRNA were not completely abolished. Thus, miR-146a down-regulated IRAK-1 and TRAF6 mRNA at the post-transcriptional level completely consistent with a previous report (11). Krutzfeldt et al. (34, 35) introduced the usage of chemically modified miRNA inhibitor known as antagomirs to define the biological functions of miRNAs. In this report miR-146a knockdown in THP-1 cells resulted in recovery from LPS tolerance and restored TNF-α secretion in response to LPS challenge (Fig. 6B). Thus, the findings fully support the dominant role of miR-146a in LPS tolerance compared with other negative regulators previously suggested as discussed above.
miR-146a Expression in LPS Stimulated Cells Contributes to Cell Survival
LPS is likely the most potent natural danger known to evoke the immune system with a systemic production of pro-inflammatory cytokines and chemokines, which recruit and activate immune cells leading to subsequent elimination of the putative infectious agent (36). More importantly, the rapid induction of cytokines such as TNF-α, IL-1, and IL-6 is indispensable for mounting the innate response and subsequent robust adaptive immune response to defend against the first invading pathogens. Although cytokine production is important for the efficient control of growth and dissemination of invading pathogens, overproduction of these cytokines is harmful for the host, because it can be detrimental to the host physiological functions and may lead to multiple organ failure and death, a condition known as the septic shock syndrome (37). Thus, cells of the innate immune system must employ constantly a multilayered control mechanism to maintain innate immunity functional and inflammation in check. For example, in the in vivo model of LPS tolerance, rats primed with a low dose of LPS (2 μg/100 g of body weight) survived a subsequent high dose of LPS challenge (1000 μg/100 g of body weight), which is known to be 100% lethal to naive rats (38). In the present study, THP-1 showed a coordinated control mechanism in the early intense production of TNF-α at the initial stage of the LPS response, and then the level of TNF-α was gradually decrease; the magnitude and timing of this response is consistent with a published study on human mononuclear cells (39). These new data demonstrating the role of miR-146a in controlling LPS tolerance confirm it as a key factor that keeps the innate immune system in balance toward cell survival.
Implication of Endotoxin Tolerance in Innate Immunity
Up-regulated miR-146a in TLR4-mediated endotoxin tolerance reported in this study is likely to affect other pattern recognition receptor activity in innate immunity. Taganov et al. (11) observed elevated miR-146a expression in response to TLR-2, -4, or -5 stimulation by bacterial and fungal components or following exposure to TNF-α or IL-1β. On the other hand miR-146a expression was not increased through activation of TLR-3, -7, or -9 in responses to viral or bacterial nucleic acids (11). Recently, Hou et al. (40) observed overexpression of miR-146a in response to vesicular stomatitis virus infection in a mouse infection model. In their study, miR-146a was shown to regulate type 1 interferon production by inhibiting RIG-I signaling molecules IRAK-1, IRAK-2, and TRAF-6 (40). Interestingly, another report by Tang et al. (25) showed the down-regulation of miR-146a in a subset of human lupus patients with elevated expression of type 1 interferon. In any case, TNF-α can be produced through activation of all TLRs, except TLR3, and all these pathways involve the miR-146a target molecules TRAF6 and IRAK-1 adaptor kinases (41). Hedl et al. (42) showed chronic stimulation of Nod2 caused down-regulation of IRAK-1 activation and rendered intestinal macrophages tolerant to muramyl dipeptide. Muramyl dipeptide activates the NF-κB pathway via the same intermediates IRAK-1 and TRAF6, and thus LPS induction of miR-146a is likely to induce cross-tolerance to Nod2 pathway. Therefore, endotoxin tolerance associated with up-regulated miR-146a (NF-κB driven) may have a broader role involved in regulating TNF-α, a key player of the cytokine network, induced by a number of pattern recognition receptors indicated in innate immunity.
Innate immune response to the invading microorganism is influenced by miR-146a. In A549 epithelial cells, Perry et al. (26) reported the IL-1β-induced release of IL-8 and RANTES (regulated on activation normal T cell expressed and secreted) that were negatively regulated by the up-regulation of miR-146a. Similarly, in LPS-treated THP-1 cells, miR-146a shows unique up-regulation that has also been observed to be responsible to cause LPS tolerance to a wide range of LPS, including S. enterica in this study. Interestingly, Cavaillon et al. (43) found that LPS-tolerant mice were significantly resistant to S. enterica or Cryptococcus neoformans infection for >2 months. Thus, overexpression of miR-146a associated with endotoxin tolerance may have important consequences in host innate immunity responding to a wide range of bacterial infection. More extensive studies are needed to fully explore the complete role of this miRNA especially in terms of its half-life in LPS-tolerant mice. It is known that, in response to LPS challenge, endotoxin-tolerant monocytic cells show reduced production of IL-1α, IL-1β, and IL-6, along with TNF-α (18). Up-regulation of miR-146a in certain inflammatory conditions such as rheumatoid arthritis and sometimes in cancer cells has been observed, although its function under those circumstances is still unclear (27, 44, 45).
In summary, multiple lines of evidence reported here provide the categorical role of miR-146a in endotoxin tolerance. This miRNA is highly up-regulated in tolerized cells and acts as a tuning mechanism to prevent an overstimulated inflammatory state. It is interesting to speculate that modulating the level of miR-146a can be used in therapeutic intervention for inflammation and protecting against sepsis.
This work was supported in part by National Institutes of Health (NIH) Grant AI47859. This work was also supported by a grant from the Lupus Research Institute.
- TLR
- Toll-like receptor
- IRAK-1
- IL-1 receptor-associated kinase 1
- LPS
- lipopolysaccharide
- miRNA
- microRNA
- pre-miR
- precursor miRNA
- TRAF6
- TNF receptor-associated factor 6
- TNF-α
- tumor necrosis factor-α
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline
- qRT
- quantitative reverse transcription.
REFERENCES
- 1.Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 2.Shishodia S., Aggarwal B. B. (2002) J. Biochem. Mol. Biol. 35, 28–40 [DOI] [PubMed] [Google Scholar]
- 3.McCall C. E., Grosso-Wilmoth L. M., LaRue K., Guzman R. N., Cousart S. L. (1993) J. Clin. Invest. 91, 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beeson P. B. (1947) J. Exp. Med. 86, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haas J. G., Baeuerle P. A., Riethmüller G., Ziegler-Heitbrock H. W. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 9563–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziegler-Heitbrock H. W., Blumenstein M., Käfferlein E., Kieper D., Petersmann I., Endres S., Flegel W. A., Northoff H., Riethmüller G., Haas J. G. (1992) Immunology 75, 264–268 [PMC free article] [PubMed] [Google Scholar]
- 7.Ulevitch R. J., Wolfson N., Virca G. D., Kim S., Kline L., Mathison J. C. (1989) Prog. Clin. Biol. Res. 299, 193–202 [PubMed] [Google Scholar]
- 8.Zuckerman S. H., Evans G. F., Snyder Y. M., Roeder W. D. (1989) J. Immunol. 143, 1223–1227 [PubMed] [Google Scholar]
- 9.Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 10.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 11.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- 13.Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M. A., Locati M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsay M. A. (2008) Trends Immunol. 29, 343–351 [DOI] [PubMed] [Google Scholar]
- 15.Li L., Cousart S., Hu J., McCall C. E. (2000) J. Biol. Chem. 275, 23340–23345 [DOI] [PubMed] [Google Scholar]
- 16.Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 17.Heagy W., Hansen C., Nieman K., West M. A. (2003) Shock 19, 321–327 [DOI] [PubMed] [Google Scholar]
- 18.LaRue K. E., McCall C. E. (1994) J. Exp. Med. 180, 2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacinto R., Hartung T., McCall C., Li L. (2002) J. Immunol. 168, 6136–6141 [DOI] [PubMed] [Google Scholar]
- 20.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 21.Shaffer J. P. (1986) J. Am. Stat. Assoc. 81, 826–831 [Google Scholar]
- 22.Guha M., Mackman N. (2001) Cell. Signal. 13, 85–94 [DOI] [PubMed] [Google Scholar]
- 23.Beutler B., Cerami A. (1987) N. Engl. J. Med. 316, 379–385 [DOI] [PubMed] [Google Scholar]
- 24.Mathison J. C., Virca G. D., Wolfson E., Tobias P. S., Glaser K., Ulevitch R. J. (1990) J. Clin. Invest. 85, 1108–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y., Luo X., Cui H., Ni X., Yuan M., Guo Y., Huang X., Zhou H., de Vries N., Tak P. P., Chen S., Shen N. (2009) Arthritis Rheum. 60, 1065–1075 [DOI] [PubMed] [Google Scholar]
- 26.Perry M. M., Moschos S. A., Williams A. E., Shepherd N. J., Larner-Svensson H. M., Lindsay M. A. (2008) J. Immunol. 180, 5689–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauley K. M., Satoh M., Chan A. L., Bubb M. R., Reeves W. H., Chan E. K. (2008) Arthritis Res. Ther. 10, R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomura F., Akashi S., Sakao Y., Sato S., Kawai T., Matsumoto M., Nakanishi K., Kimoto M., Miyake K., Takeda K., Akira S. (2000) J. Immunol. 164, 3476–3479 [DOI] [PubMed] [Google Scholar]
- 29.Dobrovolskaia M. A., Vogel S. N. (2002) Microbes. Infect. 4, 903–914 [DOI] [PubMed] [Google Scholar]
- 30.Medvedev A. E., Henneke P., Schromm A., Lien E., Ingalls R., Fenton M. J., Golenbock D. T., Vogel S. N. (2001) J. Immunol. 167, 2257–2267 [DOI] [PubMed] [Google Scholar]
- 31.Randow F., Syrbe U., Meisel C., Krausch D., Zuckermann H., Platzer C., Volk H. D. (1995) J. Exp. Med. 181, 1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berg D. J., Kühn R., Rajewsky K., Müller W., Menon S., Davidson N., Grünig G., Rennick D. (1995) J. Clin. Invest. 96, 2339–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphey E. D., Fang G., Varma T. K., Sherwood E. R. (2007) Shock 27, 289–295 [DOI] [PubMed] [Google Scholar]
- 34.Krützfeldt J., Kuwajima S., Braich R., Rajeev K. G., Pena J., Tuschl T., Manoharan M., Stoffel M. (2007) Nucleic Acids Res. 35, 2885–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005) Nature 438, 685–689 [DOI] [PubMed] [Google Scholar]
- 36.Mastroeni P., Clare S., Khan S., Harrison J. A., Hormaeche C. E., Okamura H., Kurimoto M., Dougan G. (1999) Infect. Immun. 67, 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beutler B., Milsark I. W., Cerami A. C. (1985) Science 229, 869–871 [DOI] [PubMed] [Google Scholar]
- 38.West M. A., Heagy W. (2002) Crit. Care Med. 30, S64–S73 [PubMed] [Google Scholar]
- 39.Lonnemann G., Endres S., Van der Meer J. W., Cannon J. G., Koch K. M., Dinarello C. A. (1989) Eur. J. Immunol. 19, 1531–1536 [DOI] [PubMed] [Google Scholar]
- 40.Hou J., Wang P., Lin L., Liu X., Ma F., An H., Wang Z., Cao X. (2009) J. Immunol. 183, 2150–2158 [DOI] [PubMed] [Google Scholar]
- 41.Krishnan J., Selvarajoo K., Tsuchiya M., Lee G., Choi S. (2007) Exp. Mol. Med. 39, 421–438 [DOI] [PubMed] [Google Scholar]
- 42.Hedl M., Li J., Cho J. H., Abraham C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19440–19445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavaillon J. M., Adrie C., Fitting C., Adib-Conquy M. (2003) J. Endotoxin. Res. 9, 101–107 [DOI] [PubMed] [Google Scholar]
- 44.Chan E. K., Satoh M., Pauley K. M. (2009) Arthritis Rheum. 60, 912–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanczyk J., Pedrioli D. M., Brentano F., Sanchez-Pernaute O., Kolling C., Gay R. E., Detmar M., Gay S., Kyburz D. (2008) Arthritis Rheum. 58, 1001–1009 [DOI] [PubMed] [Google Scholar]