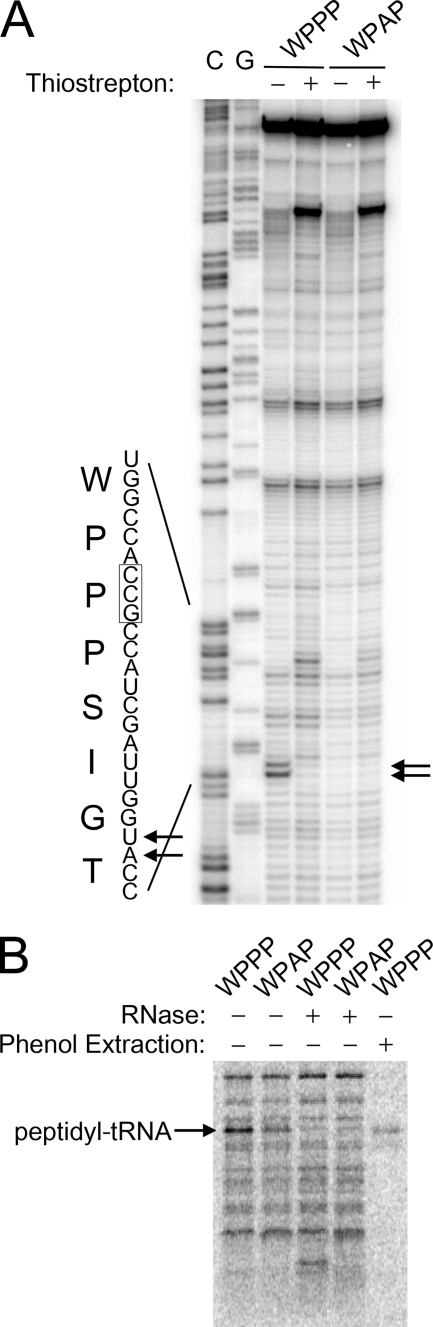
FIGURE 5.
Direct detection of stalled ribosome complexes. A, stalled ribosome complexes were formed by cell-free translation of a template encoding the FXXYXIWPPP sequence in a larger (64-mer) peptide. The non-stalling Ala mutant FXXYXIWPAP served as a negative control. The position of the ribosome was determined by reverse transcription of the mRNA template, and C and G sequencing lanes were run alongside. Thiostrepton was added in the fourth and sixth lanes to trap the ribosome in the initiation stage, demonstrating the observed toe-print signal in the third lane (marked by arrows) requires translation of the stalling site. The nucleotide and peptide sequence of the stalling site is shown at left. B, the [35S]Met-labeled products of the cell-free translation of the FXXYXIWPPP peptide or the non-stalling FXXYXIWPAP control were analyzed by Tricine-SDS-PAGE. Under these conditions, the peptidyl-tRNA linkage is not hydrolyzed during electrophoresis. The stalled peptidyl-tRNA disappears when treated with RNase (lane 3) but remains in the aqueous layer upon extraction with phenol (lane 5).