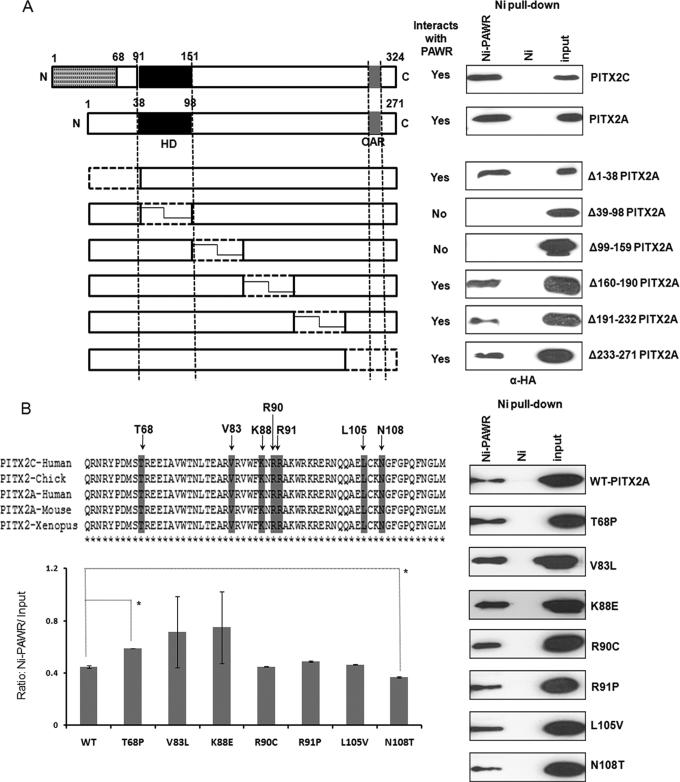
FIGURE 2.
Identification of regions in PITX2 that interacts with PAWR. A, nickel pulldown results where HTM cells transfected with six different deletion constructs (Δ1–38, Δ39–98, Δ99–159, Δ160–190, Δ191–232, and Δ233–271) named after N-terminal locations of amino acids deleted according to the A isoform of PITX2. Thus, Δ1–38 lacks first 38 amino acids of PITX2A, and so on. Subsequent nickel pulldown assays were performed with the HTM cell lysates having different deletion constructs of PITX2 and His6-tagged PAWR bound to the Ni2+-NTA beads (Ni-PAWR) or the empty beads (Ni). Bound proteins were analyzed by SDS-PAGE followed by Western blot analysis using an anti-HA antibody. N, N terminus; C, C terminus; HD, homeodomain; OAR, orthopedia and aristaless domain. B, nickel pulldown results described above but with HTM cell lysates having seven PITX2A mutant (T68P, V83L, K88E, R90C, R91P, L105V and N108T) constructs in HA-tagged pCI vector. In both cases, inputs represent 3% of the total reaction. The upper left panel of B shows the location of mutations in conserved homeodomain and the adjacent inhibitory domain, whereas the lower left panel shows Ni-PAWR/input ratio derived from quantification of immunoblots shown in the right panel. The inputs in these experiments represent 3% of the total reaction in the form of HTM cell extracts transfected with HA-tagged PITX2A wild type (WT) and mutants. Error bars indicate S.D., whereas the asterisks indicate statistical significance in the Ni-PAWR/input ratio between WT and T68P or N108T.