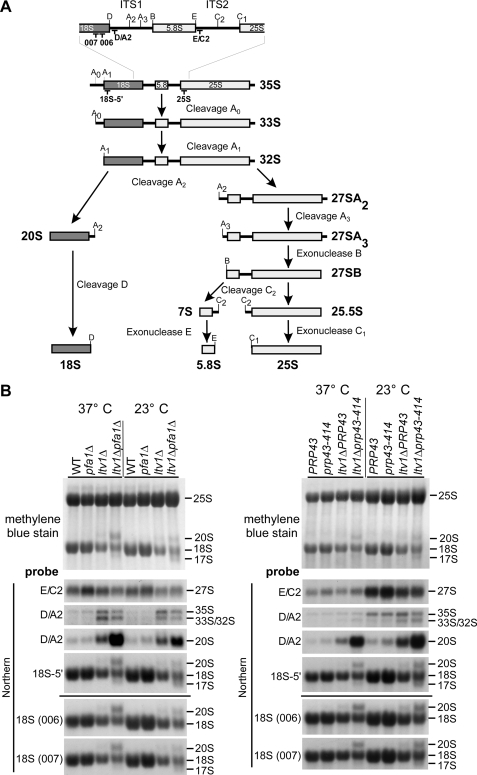
FIGURE 3.
Ltv1, Pfa1, and Prp43 are required for 20 to 18 S rRNA processing. A, simplified rRNA processing pathway in yeast. Only the major pathway for generation of the 5′-end of the 5.8 S rRNA is shown. The rRNA cleavage sites and the binding sites of the probes used for Northern blotting are indicated. ITS1 and ITS2, internal transcribed spacers 1 and 2. In the course of pre-rRNA processing, the 35 S pre-rRNA undergoes a series of endonucleolytic processing events at sites A0, A1, and A2 that lead to the separation into the 20 S and 27 S A2 pre-rRNAs. Endo- and exonucleolytic processing steps of the 27 S A2 pre-rRNA finally yield the mature 25 and 5.8 S rRNAs contained in 60 S subunits. In the cytoplasm, the final processing step in 40 S maturation takes place when the 20 S pre-rRNA is converted into the 18 S rRNA by endonucleolytic cleavage at processing site D. B, rRNA steady state levels in ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants. Cells were either grown at 37 °C to an A600 of 0.8 (37 °C) or grown at 37 °C to an A600 of 0.1, transferred to 23 °C, and further grown to an A600 of 0.8 (three cell divisions). RNA was isolated, separated by agarose gel electrophoresis, and transferred to a nylon membrane that was stained with methylene blue. rRNA processing intermediates were detected by Northern blotting using the indicated probes. WT, wild type.