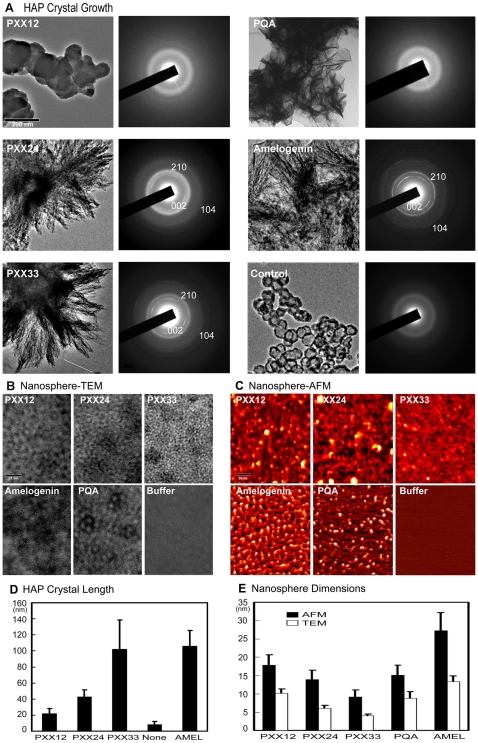
Figure 2. Hydroxyapatite crystal growth control and self-assembly of polyproline designer peptides.
(A and D) Effect of polyproline designer peptides on hydroxyapatite (HAP) crystal growth. Increasing length of designer peptides from PXX12 to PXX33 resulted in elongated HAP crystal (A). HAP crystals grown with PXX33 were similar in length to those grown with full-length amelogenin. The PQA glutamine/alanine replacement peptide resulted in thin plates unlike the needles grown with PXX repeat peptides. HAP control solutions incubated without protein formed spherical deposits. Crystal dimensions are documented in (D). Bar = 200 nm. Electron diffraction analysis of PXX24, PXX33, and amelogenin treated samples resulted in sharp and distinct reflection rings in the 002 and 210 planes and an additional faint ring in the 104 plane, similar to those found in developing enamel crystals. Control crystals and those grown in the presence of PXX12 and PQA only showed faint diffraction patterns. (B, C, and E) Effect of polyproline designer peptide length on supramolecular matrix assemblies. Both TEM and AFM analyses indicated that matrix subunit dimensions were significantly reduced with increasing designer peptide length (from PXX12 to PXX33). Full-length amelogenins formed sizable nanospheres measuring 27 nm by AFM and 13 nm by TEM in diameter. Matrix dimensions and assembly patterns of the PQA glutamine/alanine replacement peptide were significantly different from their PXX33 counterparts. Buffer control solutions did not assemble into nanosphere-like structures. All measurements in this study were statistically evaluated and displayed using standard deviation (s.d.). Bar (TEM) = 25 nm, (AFM) = 50 nm.