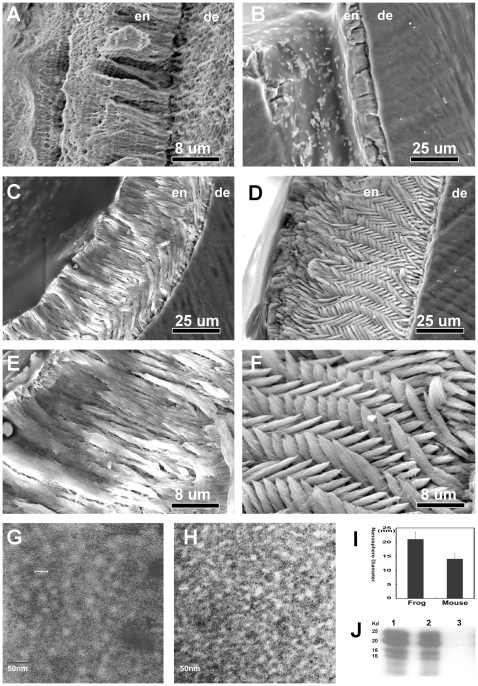
Figure 3. Differences in enamel prism formation and enamel matrix structure between molars from wild-type and frog amelogenin overexpressing mice.
Note the prism-less organization of frog enamel (A) and the complete loss of structured enamel in the amelogenin null mice (B). In these scanning electron micrographs, enamel (en) and dentin (de) were labeled for orientation purposes. Frog amelogenin overexpressing offspring crossed with amelogenin null mice (fAmel-x-null) demonstrated reduced enamel thickness (C/E versus D/F) and grossly altered enamel prism structure with fused individual crystallites, especially in the coronal half of the transgenic enamel layer (C/E versus D/F). Notably, fAmel-x-null mouse enamel lacked the regularly intercrossed prism pattern found in wild-type mouse molars (C/E versus D/F). In order to compare the effect of frog amelogenins on enamel matrix structure when compared to mouse amelogenins, enamel matrix nanosphere diameters were determined on transmission electron micrographs from the enamel matrix of developing mouse first mandibular molars (G–I). Subunits measured 20.9±2.6 nm in frog enamel chimera and 13.9±1.9 nm in their wild-type controls (G–I). Enamel matrix nanosphere dimensions in fAmel-x-null enamel exceeded those of regular mouse enamel by about 50%. (G–I) Cleavage patterns of extracted mouse enamel proteins from fAmel-x-null mice (lane 1) and wild-type mice (lane 2) were almost identical (J), while there was no amelogenin detected in amelogenin null mice (lane 3). Equal protein loads were subjected to an antibody against recombinant amelogenin for Western blotting (J).