Table 1. Over-represented GO biological process categories for modules identified from the conserved liver co-expression network among human, mouse and rat.
| M | GO Process | P | E | Background Size | Background Overlap | Module Size | Module Overlap |
| 1 | Cell-cell signaling | 2.60×10−27 | 1.70×10−23 | 5519 | 417 | 1024 | 168 |
| 2 | Translation | 7.10×10−23 | 4.70×10−19 | 5519 | 182 | 559 | 67 |
| 3 | Ribonucleoprotein biogenesis | 2.50×10−12 | 1.60×10−8 | 5519 | 111 | 527 | 37 |
| 4 | Carboxylic acid metabolic process | 4.10×10−19 | 2.70×10−15 | 5519 | 379 | 481 | 88 |
| 5 | Transcription from RNA polymerase II promoter | 5.60×10−9 | 3.70×10−5 | 5519 | 483 | 451 | 76 |
| 6 | Immune response | 2.70×10−44 | 1.70×10−40 | 5519 | 414 | 404 | 119 |
| 7 | Cell adhesion | 2.10×10−8 | 1.40×10−4 | 5519 | 493 | 112 | 30 |
| 8 | Carboxylic acid metabolic process | 1.30×10−18 | 8.40×10−15 | 5519 | 379 | 106 | 38 |
| 9 | Positive regulation of JNK cascade | 3.65×10−3 | 1.00 | 5519 | 23 | 76 | 3 |
| 10 | Cell cycle | 6.80×10−23 | 4.40×10−19 | 5519 | 254 | 74 | 31 |
| 11 | Sterol biosynthetic process | 3.20×10−30 | 2.10×10−26 | 5519 | 33 | 62 | 19 |
| 12 | Dlycerophospholipid biosynthetic process | 4.00×10−4 | 0.92 | 5519 | 34 | 24 | 3 |
| 13 | Regulation of DNA replication | 7.64×10−3 | 1.00 | 5519 | 41 | 18 | 2 |
Modules are sorted in decreasing order according to their modularity (see Materials and Methods). ‘P’ is the nominal FET p-value. ‘E’ indicates the expected false discovery rate after correcting for multiple testing (i.e. FET p-value multiplied by the total number of GO categories tested). Except modules 9, 12 and 13, all permutation adjusted p-values are 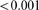 .
.
