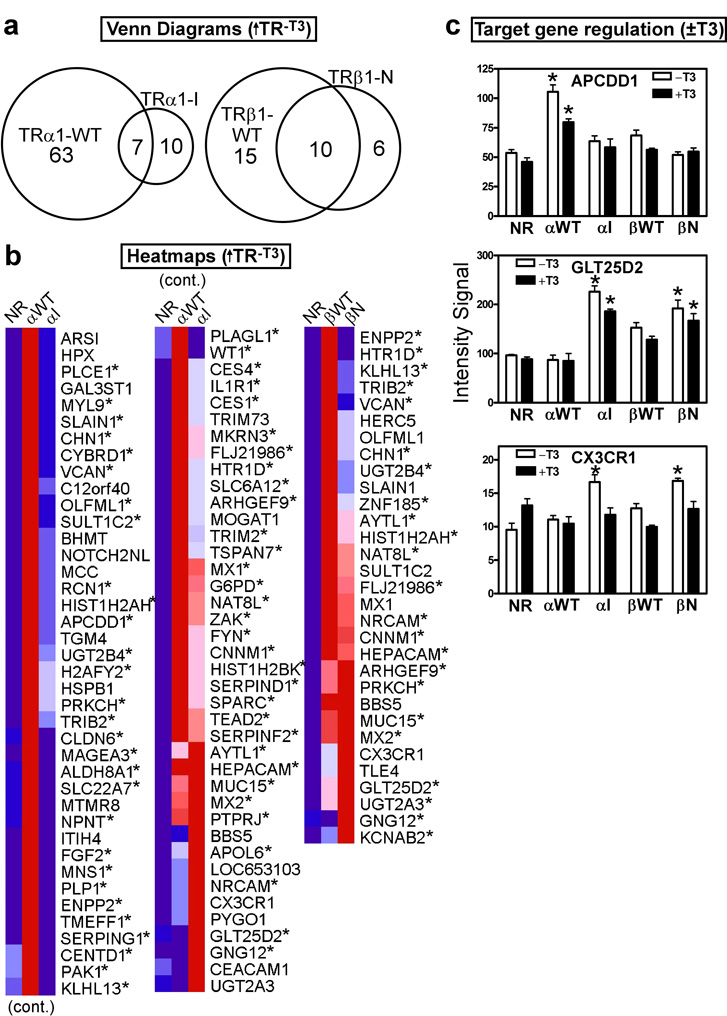
Figure 7. The mutant and wild-type TRs positively regulate distinct sets of target genes in the absence of T3.
(a) Venn diagram of gene transcripts up-regulated in each TR transformant compared to the empty vector (NR) control, all in the absence of T3. HepG2 cells transformed with the TR alleles indicated, or with the empty plasmid control, were incubated in the absence of T3 (vehicle only) for 6 hrs.; RNA was isolated, and used to probe the arrays. Transcripts up-regulated in each TR transformant compared to the empty vector control (NR) were identified using a Benjamini-Hochberg adjusted p-value of <0.05. (b) Heat map clustering of the genes from panel (a). For each gene, dark blue indicates lowest expression, dark red indicates highest expression, with intermediate values represented by lighter shades. These comparisons were minus T3; asterisks indicate genes that were also up-regulated in the presence of T3 (see Figure 4). (c) Expression levels, minus or plus T3 treatment, of representative gene transcripts from the genes identified in panel (b). Microarray intensity signal values are presented (mean + S.D., n = 3). An "*" indicates that the difference between the TR transformant and the empty vector control was significant at a P value ≤ 0.05.