Abstract
Tuftelin-interacting protein 11 (TFIP11) is a protein component of the spliceosome complex that promotes the release of the lariat-intron during late-stage splicing through a direct recruitment and interaction with DHX15/PRP43. Expression of TFIP11 is essential for cell and organismal survival. TFIP11 contains a G-patch domain, a signature motif of RNA-processing proteins that is responsible for TFIP11-DHX15 interactions. No other functional domains within TFIP11 have been described. TFIP11 is localized to distinct speckled regions within the cell nucleus, although excluded from the nucleolus. In this study sequential C-terminal deletions and mutational analyses have identified two novel protein elements in mouse TFIP11. The first domain covers amino acids 701–706 (VKDKFN) and is an atypical nuclear localization signal (NLS). The second domain is contained within amino acids 711–735 and defines TFIP11’s distinct speckled nuclear localization. The identification of a novel TFIP11 nuclear speckle targeting sequence (TFIP11-STS) suggests that this domain directly interacts with additional spliceosomal components. These data help define the mechanism of nuclear/nuclear speckle localization of the splicing factor TFIP11, with implications for it’s function.
Keywords: G-patch, lariat-intron, nuclear localization signal, RNA processing
INTRODUCTION
TFIP11 has been identified in a number of proteomic studies as a component of the nuclear spliceosome [1,2]. The spliceosome is a large complex composed of small nuclear ribonucleoproteins (snRNPs) and non-snRNP associated proteins that function together to mediate the removal of introns from pre-mRNAs. Each of the snRNPs that make up the spliceosome contains small nuclear RNAs, common Sm proteins, and its own set of specific proteins. SC35 speckles are a class of splicing-related nuclear bodies, and are identified with antibodies targeted against the splicing factor, arginine/serine-rich 2 (SFRS2) protein. TFIP11 is localized to novel subnuclear speckles that are in close proximity to, but distinct from, nuclear speckled regions characteristic of many splicing factors or snRNPs [3]. RNA interference targeting STIP (septin and tuftelin interacting protein), the C. elegans homologue of TFIP11, results in morphological abnormalities starting at about the 16-cell stage and 100% embryonic lethality [4]. The lethal phenotype can be rescued using either a Drosophila or human TFIP11 coding sequence under the control of the native C. elegans STIP promoter [4] underlining the conservation of TFIP11 function across evolution. These data highlight the fact that TFIP11 performs a non-redundant activity critical for organismal survival.
siRNA-mediated depletion of TFIP11 in HeLa cells results in a dramatic and specific accumulation of U4/U6 small nuclear ribonucleoprotein particle (snRNP) components in Cajal bodies [5], nuclear substructures that are closely associated with the import and biogenesis of many snRNPs [6]. U4/U6 snRNP accumulates in these cells indicative of impaired U4/U6.U5 snRNP assembly, implying a key role for TFIP11 in this process [6]. The yeast homologue of TFIP11, Ntr1 (Nineteen-complex related protein 1), which shows ~ 30% amino acid similarity with mammalian TFIP11s, has been shown to interact directly with Prp43, an ATP-dependent RNA helicase [7,8,9,10], thereby recruiting Prp43 to the spliceosome, a required step for the release of the lariat-intron and spliceosome disassembly in yeast [11]. Similar functional roles for TFIP11 and DHX15, the mammalian homologue of Prp43, have been described whereby TFIP11 within the U4/U6.U5 snRNP complex recruits DHX15 from the nucleoplasm, to enable the release of the lariat-intron during late-stage pre-mRNA splicing [12,13,14]. Failure of TFIP11 to recruit and interact with DHX15 results in the failure of the splicing complex to disassemble and release U2, U5, and U6 snRNPs, leading to the accumulation of post-splicing intron complexes and compromising cell behavior and survival [13]. TFIP11 contains a G-patch, which is a signature motif of many RNA-processing proteins [3,12,15]. Recent work suggests that the G-patch of TFIP11 is necessary for TFIP11-DHX15 interaction [13]. The G-patch of TFIP11 may thus serve as the DHX15 binding-domain.
Using in vitro splicing assays and cell transfection experiments we show that TFIP11 enhances splicing by promoting late-splicing events. Mutational analyses of TFIP11 define an atypical nuclear localization signal (NLS) and a sequence element directing TFIP11 to distinct nuclear speckles.
MATERIALS AND METHODS
Expression constructs
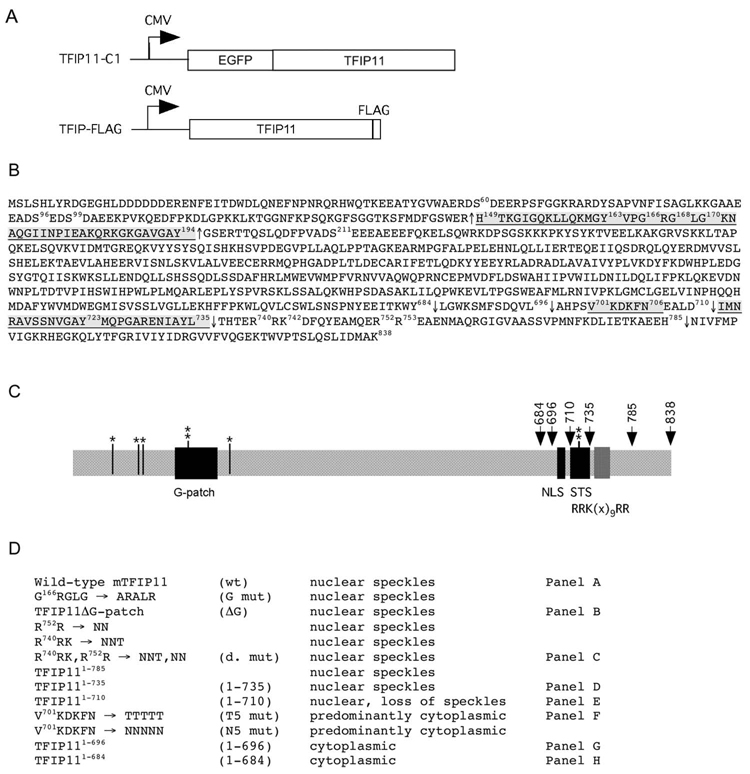
As previously reported [3], mouse TFIP11 cDNA corresponding to the entire open-reading frame (ORF) of 838 amino acids minus the initial ATG was cloned into the vector pEGFP-C1 (Clontech, Mountain View, CA) and the resulting plasmid was named TFIP11-C1 (Figure 1A). All C-terminal deletions were created in an identical manner using PCR in which the reverse primer ended as the coding sequence indicated, and this was immediately followed by a stop codon. All mutations, including the entire G-patch deletion, were performed using the GeneEditor™ in vitro Site-Directed Mutagenesis System (Promega, Madison, WI) using appropriately designed primer sets and the recommended protocols. A full-length CMV-driven TFIP11 containing 3-repeats of a FLAG-tag at the C-terminus (TFIP-FLAG) has been described previously [12] (Figure 1A). Additional TFIP11 C-terminal deletions and mutant constructs, each containing the 3X FLAG C-terminus but in all other respects equivalent to the wild-type and mutant TFIP11 fluorescent vectors prepared for immunofluorescent studies, were prepared in the backbone vector (pCMV-3Tag-8; Stratagene, La Jolla, CA) (Figure 1A) for the β-galactosidase/luciferase splicing assay described below. All constructs used in this study are illustrated and listed (Figure 1B, C and D). All PCR amplified regions were verified to be error-free by sequencing the final clones.
Figure 1. Molecular strategy for the identification of a novel NLS, and a sequence element directing TFIP11 to distinct nuclear speckles.
Panel A: schematic of the TFIP11-C1 and TFIP-FLAG constructs used in this study. Panel B: amino acid sequence for mouse TFIP11. Underlined regions correspond to the G-patch [149–194], the NLS [701–706] and the TFIP11 STS [711–735]. The ΔG mutant construct involved the removal of amino acids [149–194] and is identified by ↑, C-terminal deletion sites used for mutagenesis analysis are numbered and identified by ↓. The predicated bipartite NLS (non-functional) in TFIP11 is noted between amino acids 740–753 [R740RK(x)9RR]. Also noted are the known phosphorylation sites at S60, S96, S99, Y163, S211 and Y723. Panel C: schematic of the organization of TFIP11. Serine (S) and tyrosine (Y) phosphorylation sites identified (*, ** respectively). G-patch, NLS and STS regions shown, as is the [RRK(x)9RR] region. Panel D: brief summary table of all the constructs (wild-type and mutant) used for the analysis. Mutant constructs identified by panels A–H in figure 2 correspond to those identified on the right. Labeling in column 2 (panel D) is used to identify images in Figure 2, and identified in column 4 (panel D) and constructs in Figure 4.
Cell culture and transfection for confocal imaging
HEK293 cells were maintained in Dulbecco’s modification of Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal calf serum (FCS). For transient transfection assays, cells were grown on either four-well chamber slides (Lab-Tek) or 100-mm culture dishes. Cells were transfected using Lipofectamine 2000 (Invitrogen Corporation, Carlsbad, CA) as previously described [3]. Prior to imaging, cells were washed with phosphate buffered saline (PBS), fixed with 4% paraformaldehyde, counterstained with DAPI, mounted in VECTASHIELD medium (Vector Labs, Burlingame, CA). Direct fluorescence confocal microscopy was performed and images were captured as previously described [3,12,16].
Small interfering RNA transfection
Pre-annealed siRNAs were obtained from Ambion (Austin, TX). The control siRNA was negative silencer 1 (AM4611). The sequence of TFIP11 siRNA was 5’-CCUGUUAAGCAGGACGACUtt. HeLa cells were plated in 100-mm culture dishes so that they were ~50% confluent at the time of transfection. The cells were transfected with siRNA at the concentration of 100 nM using Oligofectamine (Invitrogen Corporation, Carlsbad, CA).
Recombinant protein
The full length human TFIP11 recombinant protein, synthesized in a wheat germ in vitro transcription-translation system [17], was purchased from Abnova (catalogue # H00024144-P01; Taipei City, Taiwan).
In vitro splicing assay
Nuclear extracts were prepared as described previously [18] with the following modifications: the high-salt nuclear extract was concentrated using pre-chilled mini-centricons (Amico, Microcon Ultracel YM-3, 3000 MWCO), and then the concentrated nuclear extract was transferred into Mini Dialysis Units (Pierce, Slide-A-Lyzer, 10,000 MWCO) and dialyzed against 250 ml dialysis buffer. The human β-globin minigene was transcribed using T7 RNA polymerase (Promega). The transcription and splicing assays were carried out as previously described [18]. The lariat-intron fraction is defined as lariat-intron/(lariat-intron + spliced products + unspliced RNA). The splicing assay was repeated at least three times. For reconstitution assay, 20 ng of recombinant TFIP11 was added in the splicing reaction prior to the incubation.
In vivo β-galactosidase/luciferase splicing assay
HEK293 cells were seeded at 4 × 105 cells/well in 12-well plate and grown in DMEM supplemented with 10% FCS. Cells were cotransfected with the double reporter pTN24 [19] and TFIP-FLAG [12] using Lipofectamine 2000 (Invitrogen Corporation, Carlsbad CA) as recommended by the manufacturer. For control, cells were cotransfected with pTN24 and pCMV-3Tag-8 (Stratagene, La Jolla, CA), which is the backbone vector of TFIP-FLAG. At 24 h after transfection, cells were washed twice with PBS and lysed in 100 µl of lysis buffer. β-galactosidase and luciferase were measured using Dual Light® system (Applied Biosystems). The ratio of luciferase activity to β-galactosidase activity represents the splicing efficiency. Transfected samples were measured in triplicate per data point. The experiment was repeated 3 times. Statistical analysis was performed using Student’s t-test.
Western blot analysis
Cell lysates and nuclear extracts were resolved by SDS-PAGE and transferred to Immobilon-P membrane (Millipore, Billerica, MA). The membranes were incubated with 8 TFIP11 antibody [12] and β–actin antibody (Sigma-Aldrich, St. Louis, MO). The protein-antibody complexes were visualized by enhanced chemiluminescence (Amersham Biosciences, GE Healthcare, Piscataway, NJ).
RESULTS
Identification of a TFIP11 nuclear speckle targeting sequence (TFIP11-STS); nuclear speckle localization is independent of the G-patch
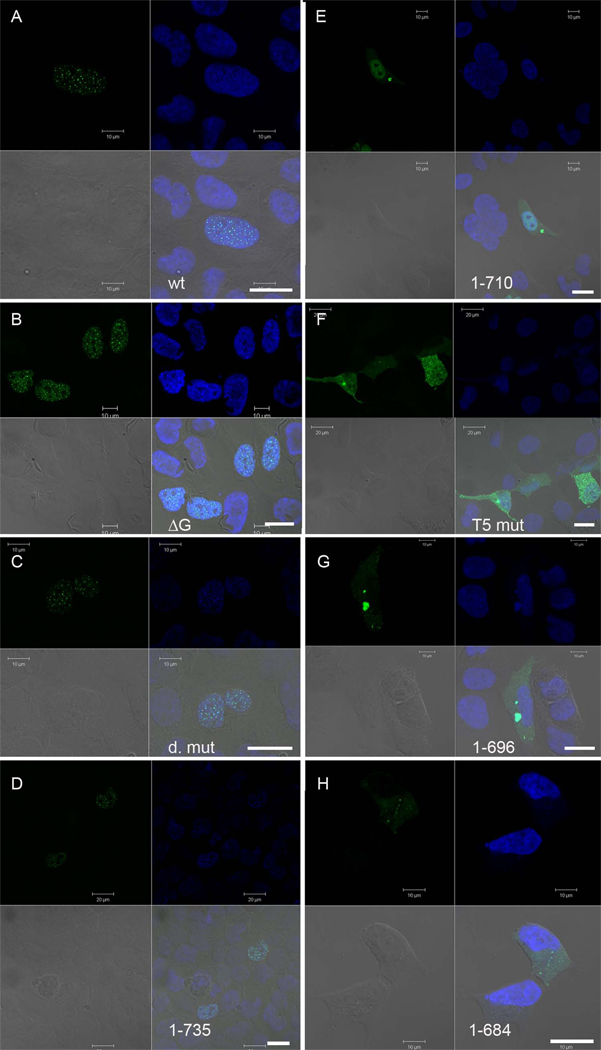
Full-length TFIP11 shows a distinct nuclear speckled location [3], but the sequences responsible have not been defined. To identify the domain necessary for this specific localization we generated a range of truncation derivatives of mouse TFIP11 fused to green fluorescent protein (GFP) (see Figure 1), and investigated their subcellular localization in transfected HEK293 cells 24h later by confocal laser scanning microscopy (Figure 2). TFIP11 amino acids 1–735 (Figure 2D) was able to target GFP to nuclear speckles to the same extent as full-length TFIP11 (Figure 2A), whereas TFIP11 amino acids 1–710 localized to the cell nucleus, but was evenly distributed throughout the nucleoplasm (Figure 2F). These results suggest clearly that the region defined by amino acids 711–735 [I711MNRAVSSNVGAYMQPGARENIAYL] is responsible for TFIP11’s distinctive nuclear speckle localization; we will refer to this region as the TFIP11 nuclear speckle targeting sequence or TFIP11-STS. The TFIP11-STS has 100% identity between mouse and human proteins.
Figure 2. Identification of a novel NLS, and a sequence element directing TFIP11 to distinct nuclear speckles using confocal microscopy.
Direct fluorescent images of TFIP11 transfected cells using either the wild-type TFIP11-C1 construct (panel A), or mutant constructs as identified in figure 1: TFIP11ΔG-patch (panel B); double mutant (panel C); TFIP111–735 (panel D); TFIP111–710 (panel E); V701KDKFN→T701TTTT (panel F); TFIP111–696 (panel G); and TFIP111–684 (panel H). Cells counterstained with DAPI, and the merged image is seen in the lower right quadrant of each panel. Also seen in the lower right quadrant is a scale bar (thick white line) for 20µm. Additional scale bars for either 10µm or 20µm, added in at the time of imaging, can be seen in each panel.
To determine whether the previously identified G-patch is involved in TFIP11 localization, we generated two G-patch TFIP11 mutants, each disrupting the highly conserved glycine residues, [G166RGLG] → [A166RALR] (Figure 1B and D) and the removal of the entire G-patch (amino acids 149–194; also referred to as ΔG-patch) (Figures 1B, C and D). Analysis of both mutant constructs demonstrated wild-type TFIP11 localization patterns (data shown for the ΔG-patch mutant, Figure 2B). These data imply strongly that the G-patch plays no significant role in nuclear/nuclear speckle localization of TFIP11.
Identification of an atypical TFIP11 nuclear localization signal (NLS)
A sequence resembling a bipartite NLS (TFIP11 amino acids 740–753; [R740RK(x)9RR]) was identified in the TFIP11 coding region, and four new constructs (see Figure 1B, C and D; [R752R]→[N752N], [R740]RK]→[N740NT], [R740RK(x)9RR]→[N740NT(x)9NN] and TFIP111–735) were generated to test functionality. Following transfections, all four constructs showed distinctive nuclear speckled localization (see Figure 2C and D; [R740RK(x)9RR]→[N740NT(x)9NN] and TFIP111–735 respectively) comparable to that of wild-type TFIP11 (Figure 2A). Clearly, the putative NLS was not essential for TFIP11 nuclear/nuclear speckle localization.
The fact that TFIP11 amino acids 1–710 conferred even distribution throughout the nucleoplasm (Figure 2E), whilst TFIP11 amino acids 1–696 mediated entirely cytoplasmic localization (Figure 2G), implied that the TFIP11 NLS is located between amino acids 697–710. This sequence [A697HPSVKDKFNEALD], again 100% conserved in human TFIP11, represents a novel region that contains several lysine residues, but mostly non-basic polar amino acids. Two point mutations [V701KDKFN]→[T701TTTT] (Figure 2F), or [N701NNNN] (data not shown) were generated in the full length TFIP11 sequence, both resulting in predominantly cytoplasmic localization of GFP; clearly, V701KDKFN is essential for nuclear localization of TFIP11.
TFIP11 is involved in spliceosome disassembly and enhances splicing activity in vivo
Several studies on budding yeast have demonstrated that Ntr1 is associated with the post-splicing excised intron complex, and that it is essential for the release of lariat-introns from the spliceosome [7,8,11]. The G-patch domain of Ntr1 has been shown to interact with Prp43 and this interaction is critical for spliceosome disassembly [8]. Like its yeast counterpart, TFIP11 interacts with the mammalian homologue of Prp43 [12,13].
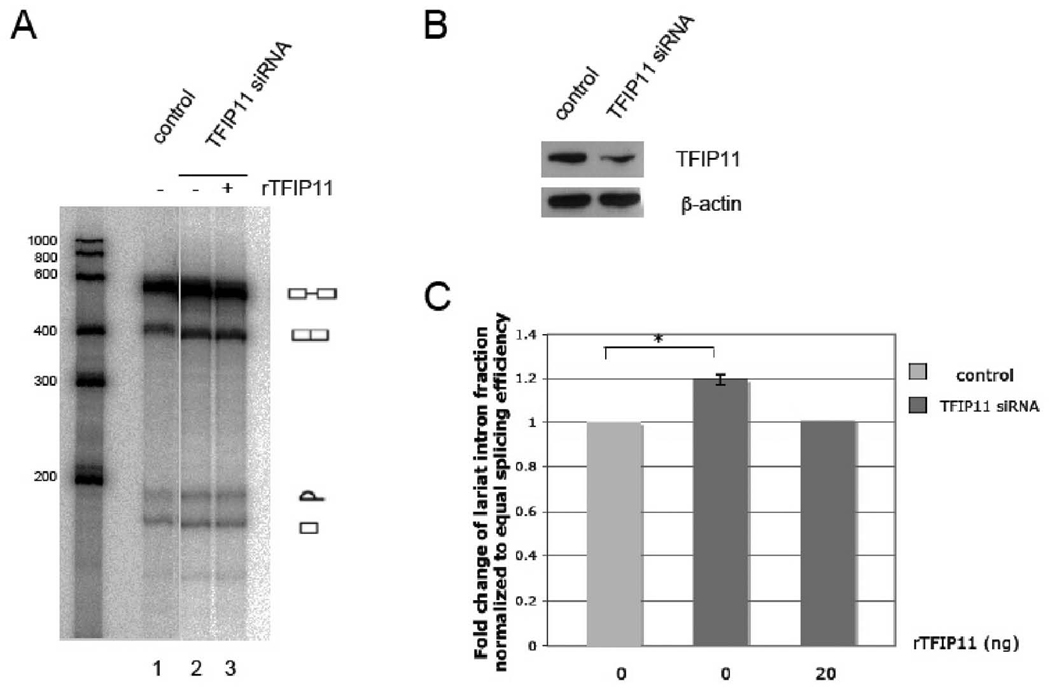
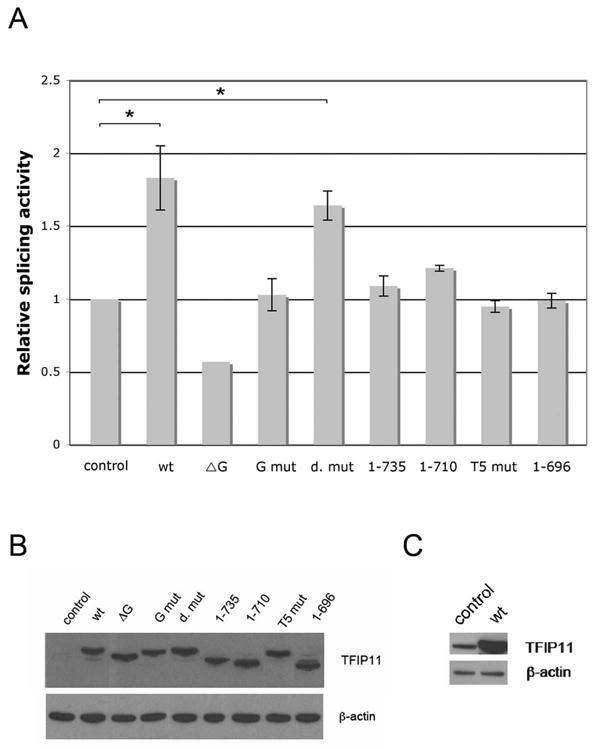
To examine the functional role of TFIP11 in pre-mRNA splicing, we performed in vitro splicing assays using nuclear extracts generated from HeLa cells either transfected with negative silencer or TFIP11 siRNA. In HeLa cells transfected with TFIP11 siRNA, TFIP11 protein was depleted approximately 70% (Figure 3B). Depletion of TFIP11 resulted in significantly increased amounts of the lariat intron (Figure 3A, lane 2). Importantly, this defect was rescued by recombinant TFIP11 in add-back experiment (Figure 3A, lane 3). These results strongly support the notion that TFIP11 is involved in the release of excised introns from spliceosomal complexes. These observations also suggest that TFIP11 promotes pre-mRNA splicing. To test this hypothesis, we carried out TFIP11 overexpression assays using a cell culture double-reporter splicing assay in which luciferase is expressed only after intron removal [19]. HEK cells were cotransfected with the reporter and an expression construct encoding TFIP-FLAG. Splicing efficiency was then quantitated relative to that of cells cotransfected with reporter and empty FLAG vector (pCMV-3Tag-8). In support of the hypothesis that wild-type TFIP11 promotes efficient pre-mRNA splicing, we observe that TFIP11 overexpression results in an ~ 1.7–1.8 fold activation of the splicing efficiency (p < 0.05) (Figure 4A). This 1-7-1.8 fold greater splicing efficiency was unchanged when the putative NLS was mutated ([R740RK(x)9RR]→[N740NT(x)9NN]; also identified as d. mut) with a p < 0.05 (Figure 4A). With the exception of the deleted G-patch mutant TFIP11 vector (ΔG), all other TFIP11 mutant vectors tested restored the splicing efficiency to baseline (or control) levels (Figure 4A). The G-patch deleted mutant vector (ΔG) appears to have a dominant-negative impact on splicing activity decreasing splicing efficiency by ~ 50% (to that seen in the control).
Figure 3. Accumulation of lariat intron in TFIP11-depleted nuclear extract.
A, Human β-globin pre-mRNA was incubated with HeLa nuclear extracts. For reconstitution assay, 20 ng of recombinant TFIP11 was added in the splicing reaction. B, TFIP11 expression in nuclear extracts of HeLa cells transfected with negative silencer or TFIP11 siRNA was detected by immunoblot using rabbit-generated antibody against TFIP11 and β–actin included as control. C, Quantitation of the lariat intron fraction. Error bar represents the standard error of mean. Statistically significant comparison data indicated by *, p < 0.05.
Figure 4. TFIP11 stimulates splicing activity in vivo.
A, TFIP-FLAG, pCMV-3Tag-8 (control) and a series of mutated TFIP11 constructs containing a C-terminus FLAG tag were cotransfected with double reporter plasmid pTN24 in HEK293 cells. The ratio of luciferase to β-galactosidase acitivities were calculated as relative splicing activity with the activity of the empty vector set at 1. The results shown here (with the exception of the ΔG vector where the experiment was repeated only once) represent three independent experiments in which three wells of cells were transfected and each cell lysate was measured in triplicate. Standard error of mean shown. Statistically significant comparison data for the wt and d. mut vectors, when compared to control, indicated by *, p < 0.05. B, Immunoblot analysis of cell lysates from pCMV-3Tag-8 (control), TFIP-FLAG (wt) and mutated TFIP11 constructs (ΔG, G mut, d. mut, 1–735, 1–710, T5 mut and 1–696) transfected cells using rabbit-generated antibody against TFIP11 and β–actin included as control. C, The band for the control (endogenous TFIP11; left column), compared to transfected TFIP-FLAG (wt; right column), became apparent when film exposed for a longer time.
Based on these functional assays and previous results, we conclude that TFIP11 is involved in maintaining efficient intron removal by accelerating the recycling of functional snRNPs.
DISCUSSION
We demonstrate here for the first time the splicing activity of TFIP11 both in vitro and in vivo, and importantly, define the sequences within TFIP11 that determine its nuclear/nuclear speckle localization critical to its splicing function. Prior to this study, the only feature of TFIP11 that had been examined was the G-patch domain, a signature motif for RNA-processing proteins; intriguingly, although the G-patch domain is usually found in combination with other RNA-binding motifs in other proteins, this is not the case for TFIP11, implying a distinctive function for TFIP11, consistent with the effects of anti-TFIP11 siRNA on cell function/viability [4]. Splicing factors generally have a characteristic speckled location within the cell nucleus, where molecular components of the spliceosome reside, moving to active sites of transcription/pre-mRNA splicing as required, but analysis of the sequences responsible for nuclear speckle localization have not been performed. Previously we showed that TFIP11 resides in close proximity to nuclear speckles [3]. Yoshimoto and colleagues characterized TFIP11 as a component of the post-splicing lariat-intron complex [13], and a number of reports indicate that TFIP11, like its yeast homologue Ntr1, recruits DHX15 (Prp43) from the nucleoplasm to allow the subsequent release of the lariat-introns from the splicing complex [8,9,11,12]. Recently, Stanek and colleagues have shown that siRNA targeted depletion of TFIP11 results in accumulation of U4/U6 snRNP in Cajal bodies, suggesting TFIP11’s role in snRNP recycling [5]. We show that depletion of TFIP11 results in accumulation of lariat intron. The data here complement the previous studies on the role of human TFIP11 during spliceosome disassembly, and further solidify the notion that TFIP11 is involved in the release of lariat intron during late stages of the pre-mRNA splicing pathway. Using an in vivo double-reporter splicing assay [19], we show directly that TFIP11 upregulation increases the splicing activity. This increase in splicing activity is likely related to the more efficient recycling of splicing factors.
Mutational analyses of TFIP11, using either the T5 mut or N5 mut vectors (Figure 1, panel D) enabled the identification of an atypical NLS and a region that alone defines a distinct nuclear speckle localization of TFIP11 (Figure 2, panel F). In addition, mutation of the atypical NLS (T5 mut; Figure 4A) was sufficient to eliminate any splicing enhancement, over baseline (Figure 4A, control), using the double-reporter splicing assay. C-terminal deletion analyses of TFIP11 enabled the identification of TFIP11-STS. The TFIP11-STS may be a region that interacts directly with a structural component (protein or RNA) of the TFIP11 storage site. However, it remains unknown how the structural integrity of subnuclear organelles devoid of enclosed membrane is maintained in highly organized nuclei.
TFIP11 is known to undergo post-translational modification, phosphoproteomic analyses showing that that at-least 4 serine and 2 tyrosine residues (Figure 1B and C) undergo phosphorylation modification [20,21,22,23]. This is a reminiscent to the activation of SR proteins, a family of non-snRNP splicing factors. SR proteins have been shown to be regulated by phosphorylation, which alters their subcellular localization and affects their ability to interact with RNA/proteins [24]. Similarly, phosphorylation events may be critical for nuclear transport of TFIP11, TFIP11-DHX15 interactions, or additional TFIP11-protein or TFIP11-RNA interactions.
TFIP11 is critical for organismal survival, highlighting the importance of a correctly functioning pre-mRNA splicing complex. To date no disease process has been identified for a mutated TFIP11 gene locus, but aberrant splicing activities are noted in many cancers [25,26], and the molecular mechanisms responsible may relate to key splicing factor activities being adversely impacted by specific mutations. Previously, we showed that TFIP11 interacts with cyclin L1 (CCNL1) and Ewing’s sarcoma protein (EWSR1), both of which have been previously associated with pre-mRNA splicing events [16]. It is intriguing that TFIP11 may participate in multiple cellular activities as diverse as pre-mRNA splicing, cell cycle activity and tumorigenesis.
CONCLUSION
In conclusion, TFIP11 is a protein component of the spliceosomal complex and is directly involved in the disassembly of snRNPs associated with lariat-introns during late-stage splicing events. Using fluorescent microscopy, and a truncation/mutagenic approach, two regions within TFIP11 have been identified that define its unique nuclear localization properties; the NLS and the TFIP11-STS. TFIP11’s G-patch has no role in determining TFIP11 nuclear localization but it is essential for interactions with DHX15.
Acknowledgments
We are grateful to Dr. Md.Talat Nasim for providing the double reporter vector pTN24. We are also thankful to members of the Hertel (University of California, Irvine) and Paine (University of Southern California) laboratories for their expertise and support. We would like to thank Michelle Mac Veigh for helping with the confocal microscopy. This work was supported by NIDCR grant DE014867 (M.L.P.), and by NIGMS grant GM062287 (K.J.H.) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 2.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2002;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 3.Wen X, Lei YP, Zhou YL, Okamoto CT, Snead ML, Paine ML. Structural organization and cellular localization of tuftelin-interacting protein 11 (TFIP11) Cell. Mol. Life Sci. 2005;62:1038–1046. doi: 10.1007/s00018-005-4547-z. [DOI] [PubMed] [Google Scholar]
- 4.Ji Q, Huang CH, Peng J, Hashmi S, Ye T, Chen Y. Characterization of STIP, a multi-domain nuclear protein, highly conserved in metazoans, and essential for embryogenesis in Caenorhabditis elegans. Exp. Cell Res. 2007;313:1460–1472. doi: 10.1016/j.yexcr.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol. Biol. Cell. 2008;19:2534–2543. doi: 10.1091/mbc.E07-12-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris GE. The Cajal body. Biochim. Biophys. Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol. Cell Biol. 2006;26:6016–6023. doi: 10.1128/MCB.02347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, Yeh FL, Cheng SC. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell Biol. 2007;27:8027–8037. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandit S, Lynn B, Rymond BC. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. USA. 2006;103:13700–13705. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen X, Tannukit S, Paine ML. TFIP11 interacts with mDEAH9, an RNA helicase involved in spliceosome disassembly. Int. J. Mol. Sci. 2008;9:2105–2113. doi: 10.3390/ijms9112105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshimoto R, Kataoka N, Okawa K, Ohno M. Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res. 2009:891–902. doi: 10.1093/nar/gkn1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tannukit S, Wen X, Wang H, Paine ML. TFIP11, CCNL1 and EWSR1 protein-protein interactions, and their nuclear localization. Int. J. Mol. Sci. 2008;9:1504–1514. doi: 10.3390/ijms9081504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madin K, Sawasaki T, Ogasawara T, Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. USA. 2000;97:559–564. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks MJ, Lam BJ, Hertel KJ. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods. 2005;37:306–313. doi: 10.1016/j.ymeth.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Nasim MT, Eperon IC. A double-reporter splicing assay for determining splicing efficiency in mammalian cells. Nat. Protoc. 2006;1:1022–1028. doi: 10.1038/nprot.2006.148. [DOI] [PubMed] [Google Scholar]
- 20.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeakley JM, Tronchere H, Olesen J, Dyck JA, Wang HY, Fu XD. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettigrew CA, Brown MA. Pre-mRNA splicing aberrations and cancer. Front. Biosci. 2008;13:1090–1105. doi: 10.2741/2747. [DOI] [PubMed] [Google Scholar]
- 26.Xing Y. Genomic analysis of RNA alternative splicing in cancers. Front. Biosci. 2007;12:4034–4041. doi: 10.2741/2369. [DOI] [PubMed] [Google Scholar]