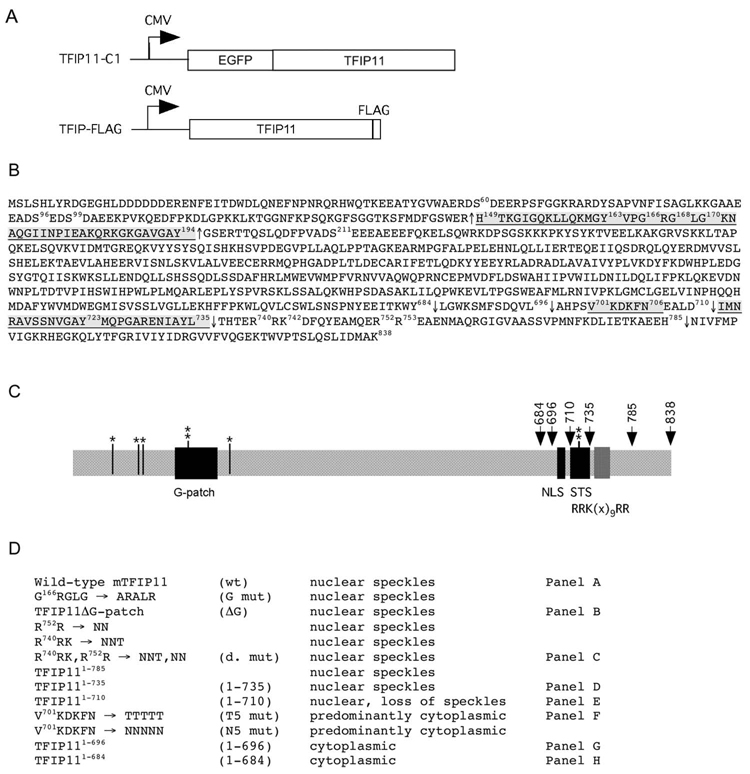
Figure 1. Molecular strategy for the identification of a novel NLS, and a sequence element directing TFIP11 to distinct nuclear speckles.
Panel A: schematic of the TFIP11-C1 and TFIP-FLAG constructs used in this study. Panel B: amino acid sequence for mouse TFIP11. Underlined regions correspond to the G-patch [149–194], the NLS [701–706] and the TFIP11 STS [711–735]. The ΔG mutant construct involved the removal of amino acids [149–194] and is identified by ↑, C-terminal deletion sites used for mutagenesis analysis are numbered and identified by ↓. The predicated bipartite NLS (non-functional) in TFIP11 is noted between amino acids 740–753 [R740RK(x)9RR]. Also noted are the known phosphorylation sites at S60, S96, S99, Y163, S211 and Y723. Panel C: schematic of the organization of TFIP11. Serine (S) and tyrosine (Y) phosphorylation sites identified (*, ** respectively). G-patch, NLS and STS regions shown, as is the [RRK(x)9RR] region. Panel D: brief summary table of all the constructs (wild-type and mutant) used for the analysis. Mutant constructs identified by panels A–H in figure 2 correspond to those identified on the right. Labeling in column 2 (panel D) is used to identify images in Figure 2, and identified in column 4 (panel D) and constructs in Figure 4.