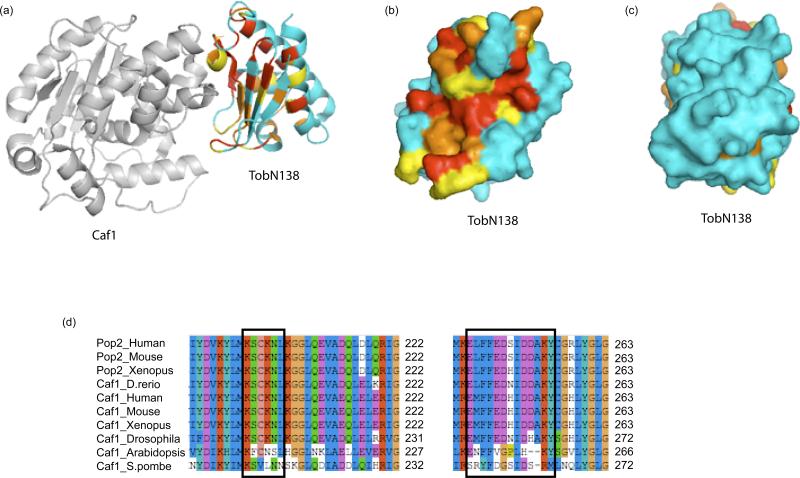
Figure 3.
The APRO domain is a conserved adaptor binding the Caf1 deadenylase.
(a) Structure of the CAF1-TOBN138 complex shown as ribbon diagram (PDB entry 2D5R, [12]). CAF1 is shown in gray. For TOBN138, invariant residues are shown in red, well-conserved residues in orange, moderately conserved residues in yellow and non-conserved residues in blue. The degree of conservation is taken from a ClustalW multiple sequence alignment of vertebrate APRO domains (shown in supplemental data). All structural images were prepared using PyMOL (www.pymol.org).
(b) Surface representation of TOBN138 showing the surface in contact with CAF1. The coloring scheme is as in (a).
(c) Surface representation of TOBN138 showing the opposite face as compared to (b). The coloring scheme is as in (a).
(d) The CAF1 regions interacting with TOB1 are conserved only in orthologs from species encoding APRO domains. Clustal W sequence alignments of two regions of CAF1 are shown. Residues found to contact TOBN138 [12] are indicated in black boxes. Note that these residues are not conserved in Arabidopsis and S. pombe, which do not encode APRO-containing proteins in their genomes.