Abstract
Transthyretin (TTR) is an attractive candidate for use in phylogenetic analysis because it is a short, single-copy nuclear gene with regions that are highly conserved across evolutionarily-divergent organisms from Xenopus laevis to Homo sapiens. To explore its utility as a phylogenetic marker, the complete intron one region (789–805 bp) was sequenced in 22 crocodylian species. Detailed analyses of intron 1 resolved the three expected lineages, Alligatorids, Crocodylids, and Gavialids, and offered additional evidence for the utility of synapomorphic indels in elucidating higher-level phylogenetic relationships. When used in conjunction with other genetic and morphological data sets, intron 1 should be a valuable tool in the investigation of other closely-related taxa.
Keywords: TTR intron 1, Transthyretin, Mecistops cataphractus, Tomistoma schlegelii, Gavialis gangeticus, Alligatoridae, Gavialidae, Crocodylidae, Systematics
The gene encoding transthyretin (TTR) is an attractive candidate for use in phylogenetic analysis because it is a short, single-copy nuclear gene with coding regions that are highly conserved across evolutionarily-divergent organisms. The TTR gene product belongs to a group of thyroid hormone binding proteins that also includes thyroxine-binding globulin and albumin. The gene spans ~6.9 kilobases and consists of 4 exons and 3 introns (Power et al., 2000). Sequence identity between diverse species is relatively high, and the hormone binding site is highly conserved across almost all vertebrate taxa. For example, sequence comparisons of TTR between eutherians, marsupials, birds, and lizards show 65–85% similarity between these diverse groups (Power et al., 2000). Although a few investigators have used the sequence of TTR mRNA to discern phylogenetic relationships, most studies have focused on the evolution of transthyretin itself (Hennebrey et al., 2006; Prapunpoj et al., 2002). Moreover, the use of mRNA for phylogenetic analysis introduces the additional difficulty of acquiring tissue samples. This becomes even more problematic with lineages that are threatened or endangered. A better strategy targets the first intron of the TTR gene. It is well-suited for sequencing because its short length is conserved across a wide variety of taxa. Additionally, the flanking coding regions are relatively long and have sections that are also highly conserved, facilitating primer design for comparison of diverse species. Two initial efforts to use TTR intron 1 for analysis of limited mammalian lineages have been promising (Steiner et al., 2005; Walton et al., 2000), but they focused on specimens that were highly divergent (21% and 31% respectively). It remains to be seen if this marker will prove to be useful for more closely-related taxa.
The extant crocodylians may provide an excellent group for testing the reliability of this potential marker among closely-related species. There are extensive molecular and morphological data sets available for comparison. Moreover, an extensive fossil record implies that some species diverged from other groups approximately 84 mya (Brochu, 2003). On the other hand, there also appear to be species in these lineages that have diverged recently (~5mya). Historically, crocodylians have been divided among three different lineages (Alligatoridae, Crocodylidae, and Gavialis). All three groups include extinct relatives that are not placed within the crown clades comprising stem alligatoroids, crocodyloids and gavialoids. Nevertheless, their similarities have left some controversy about the details of their relationships. For example, the false gharial (Tomistoma schlegelii), has traditionally been placed within Crocodylidae (Brochu, 1999), but most molecular analyses imply a Gavialid relationship (Densmore, 1983; Densmore and Dessauer, 1984; Gatesey et al., 2003; Harshman et al., 2003). It therefore seemed likely that detailed comparisons of TTR intron 1 among the crocodylians would both establish the utility of this marker for phylogenetic analysis and address some ambiguities among the relationships within this widespread group of reptiles.
MATERIALS AND METHODS
Blood collection and DNA Extraction
Whole blood of various crocodylians was collected from the dorsal postcranial sinus (Bayliss, 1987) and was used as the source of DNA for this study. These samples were a generous gift from Dr. Llewellyn D. Densmore, III. DNA extractions from all the species included in the study were performed using a PureGene DNA extraction kit (Minneapolis, MN) or MoBio UltraClean DNA BloodSpin kit (Calrsbad, CA) with minor modifications to the manufacturers’ protocols.
Amplification of Nuclear Genes
Polymerase chain reaction (PCR) amplifications of the targeted TTR gene were performed using gene-specific forward primer 5′-GGCTTTTCATTCTATGCTTCTCG-3′ located near the 3′ terminus of exon1 developed by Prapunpoj et al. (2002) and a reverse primer that I designed based on a region located downstream of the 5′ end of exon 2, 5′-CTTGCCAGTCTCCATCTGAAG-3′. Each reaction consisted of Failsafe PCR System (Epicentre Biotechnologies, Maddison, WI) with 1μl of each primer (10μM), 0.5 mu;l of Failsafe PCR Enzyme mix (1.25U), and water to a final volume of 25 mu;l, to which I added 25 mu;l of Premix F containing MgCl2 (~5mM), dNTP (400 mu;M each) Tris-HCl (100mM), and KCl (100mM). Amplification began with an initial denaturing step of 95°C for 5 minutes and thirty cycles were then performed with the following parameters: 40-second denaturation at 95°C; 40-second annealing at 55°C; and a 90-second extension at 72°C, all conducted in an Eppendorf Mastercycler gradient thermocycler (Brinkmann Instruments Inc., Westbury, NY). Amplification ended with a 7-minute extension step of 72°C followed by a 4°C hold. The amplified products were purified using DNA Clean and Concentrator-5 (Zymo Research, Orange, CA) following the manufacturer’s protocol and a cycle sequencing reaction was performed using the previously noted primers. After a final purification step using ZR DNA Sequencing Clean-up kit (Zymo Research, Orange, CA), the PCR products were sequenced in an ABI 3130 automated sequencer (Perkin Elmer, Foster City, CA), with ABI Big Dye Chemistry v. 3.1 (Perkin Elmer, Foster City, CA). The resulting sequence information from 22 species has been deposited in GenBank with the accession numbers, GQ496361-GQ496382.
Data Analyses
Sequences were aligned using Align X in Vector NTI Advance Suite software version 10.0 (Invitrogen Inc., 2006) and verified manually for obvious sequence misreads. Alligator mississippiensis was used as the outgroup taxon in all analyses. Gene sequences were analyzed using maximum parsimony and maximum-likelihood using PAUP v. 4.0b10 (Swofford, 2002), and Bayesian methods using Mr. BAYES v3.0 (Huelsenbeck and Ronquist, 2001). Sequence divergence values were calculated using uncorrected pair-wise values and k2p parameters. In maximum-likelihood analyses, Modeltest (Posada and Crandall, 1998) determined the most appropriate evolutionary model to be HKY85 (Hasegawa et al., 1985) using base frequencies estimated by PAUP (A=0.32067, C=0.1623, G=0.16413, T=0.35286) and a heuristic search was performed with 100 bootstrap replicates.
In Bayesian analyses, MrModeltest (Nylander, 2004) determined the most appropriate evolutionary model to be GTR+I+G (Laneve et al., 1984). To evaluate the parameters used, multiple independent Metropolis-coupled MCMC runs from random starting points were performed including a cold chain followed by five incrementally heated chains. A starting tree was chosen at random and 1.0 × 107 generations were run with sampling every 100 generations using the most appropriate model of evolution for the dataset. In all searches, stationarity of the Markov chain was determined as the point when sampled log-likelihood values plotted against generation time reached a stable value. Additionally, the on-line application AWTY (Wilgenbush et al.; 2004) and its cumulative function to verify post burn-in values did not vary directionally over time. A burn-in of 5000 trees was then set producing 95001 sample points. The resulting trees were used to generate a majority consensus tree with posterior probability values. Nodes with values of 85 to 89 were considered to have low support, 90 to 94 to have moderate support and nodes greater than 95 to be highly supported (Huelsenbeck and Ronquist, 2001).
RESULTS
To explore the utility of the transthyretin gene in systematic analysis, I used the crocodylians as a model phylogenetic group. Approximately 980 base pairs spanning TTR intron 1 and sections of exon 1 and exon 2 were sequenced for twenty-two individuals representing every crocodylian species except Crocodylus novaeguineae. This enabled me to compare the complete 789–805 bp introns for all species noted in the study. Initially, both primers designed by Prapunpoj et al. (2002) were used to sequence the first animals in this study. However, these did not provide the length needed to sequence the entire intron without cloning. Consequently, a new reverse primer was designed based on a conserved region 76 base pairs further downstream so that the fragment could be sequenced directly from the PCR product. Intrageneric variation was moderate, with most occurring in alligatorids. The crocodylids were more conserved with the exception of Osteolaemus tetraspis and Mecistops cataphractus. These species were differentiated from the others by several unique transitions and transversions. Of the 789–805 base pairs examined, there were 718 constant sites, 32 parsimony uninformative sites, and 55 parsimony informative sites.
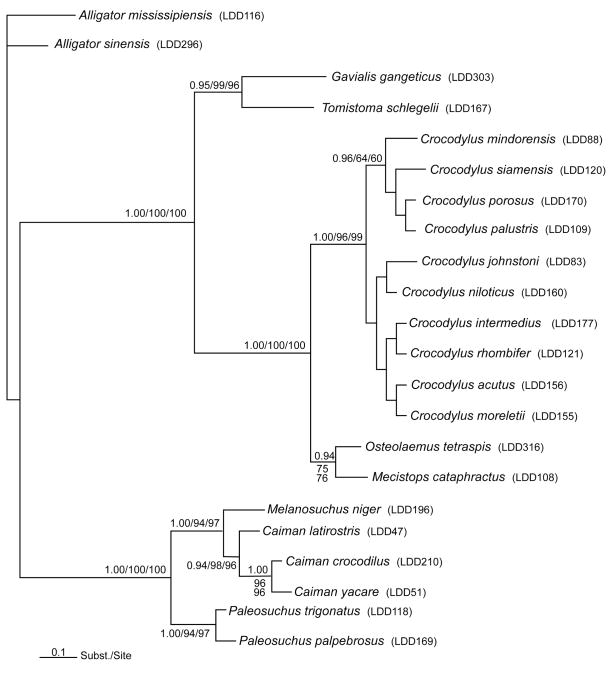
Maximum-likelihood (ML), maximum parsimony (MP), and Bayesian analyses produced phylogenetic trees with essentially identical topologies. The only major difference was that the MP and ML had more difficulty elucidating Crocodylus relationships and most were placed into an unresolved polytomy. Bayesian posterior probability values provide strong support at most major nodes (i.e., at the generic level, Fig. 1), but not among species in the Crocodylus genus. As occurred with the MP and ML analysis, however, the dataset differentiated between the “Old World” and “New World” clades. All New World true crocodiles (C. acutus, C. intermedius, C. moreletii, and C. rhombifer) were united in a single clade. The Old World true crocodiles (C. mindorensis, C. palustris, C. porosus, and C. siamensis) also formed a clade with the exception of C. niloticus and C. johnstoni, which formed their own clade closer to the New World group (Fig. 1). Mecistops cataphractus maintained a sister-taxon relationship with Osteolaemus and formed a clade separate from all other Crocodylus with high support values (Fig. 1). Indeed, the support for classifying Mecistops and Osteolaemus as sister taxa is higher than some of the data sets that originally separated Mecistops from Crocodylus (McAliley et al., 2006). The false gharial (Tomistoma schlegelii) formed a clade with the true gharial (Gavialis gangeticus) with high support values in all three analyses. All phylogenetic analyses strongly support the clades formed within the caiman complex (Caiman spp., Melanosuchus niger and Paleosuchus spp.) Melanosuchus formed a clade with the members of the Caiman genus, and Paleosuchus trigonatus and P. palpebrosus were in their own clade. Finally, the Chinese alligator (Alligator sinensis) formed the sister taxon to the American alligator (Alligator mississippiensis), which was used as the outgroup in all three analyses.
Figure 1.
Consensus Bayesian tree illustrating the relationships of crocodylians using the evolutionary model of GTR+I+G (Lanave et al., 1984) and sequences from the nuclear gene TTR intron 1. Starting tree was chosen at random and 1.0 × 107 generations run with sampling every 100 generations and a burnin of 5000 resulting in 95001 sample points. Values near nodes are Bayesian posterior probability values followed by maximum-likelihood and maximum-parsimony bootstrap values. Nodes with values below 50% were not shown.
Genetic distance values using uncorrected pair-wise values or K2p parameters were virtually identical. As such, only the uncorrected pair-wise value is shown in the table (Table 1). The values ranged from less than 0.3% when comparing species within Crocodylus to a high of 6.85% between members of Crocodylus and the caimans. It is noteworthy that Mecistops cataphractus had a consistently higher level of divergence than Osteolaemus when compared to the remaining members of Crocodylus. Mecistops was found to be only 0.43% divergent from Osteolaemus and displayed 1.0% percent divergence to the closest Crocodylus. The false gharial was 1.55% divergent from the true gharial and both of these animals were 2.8% divergent from most of Crocodylus.
Table 1.
Uncorrected pair-wise genetic distance values for the nuclear gene TTR intron 1
| Crocodylians | TTR Intron One (%) |
|---|---|
| Gavialis to A. mississippiensis | 3.95% |
| Gavialis to Crocodylus spp. | 2.90% |
| Gavialis to Tomistoma | 1.55% |
| Osteolaemus to C. niloticus | 0.89% |
| Mecistops to C. niloticus | 1.00% |
| Mecistops to Osteolaemus | 0.43% |
| A. mississippiensis to C. niloticus | 4.33% |
| C. mindorensis to Caiman yacare | 6.85% |
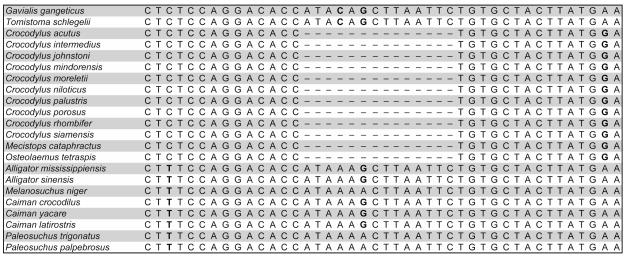
There were synapomorphic indels present within the data set. The most notable was a 14-bp indel in Gavialis gangeticus, Tomistoma schlegelii and all alligatorids, which was not present in Crocodylus, Mecistops and Osteolaemus (Fig. 2).
Figure 2.
Partial sequence of the TTR intron 1 nuclear gene highlighting the 14 base-pair indel characteristics of the extant crocodylians.
DISCUSSION
The phylogenetic comparisons of TTR intron 1 corroborates several relationships within the crocodylians that have been historically supported by other molecular data (Brochu and Densmore, 2001; Ray and Densmore, 2002; Roos et al., 2007) with high support in all three analyses (ML, MP and Bayesian). Crocodylians have been extensively studied and most relationships have been discerned using morphological, as well as mitochondrial and nuclear gene comparisons. However, there are still some relationships that have not been fully resolved and most data sets have not been able to elucidate all nodes across crocodylian relationships. The last few years have seen resurgence in crocodylian systematics in an effort to find a marker that could consistently elucidate all crocodylian lineages. Most of the recent data sets have not included all of the extant crocodylians. Not having every representative can hinder the ability to discern if the marker is useful or if it is just a consequence of not having enough samples for the lineages to fall into their evolutionary relationships.
The caimans (Caiman spp., Melanosuchus niger and Paleosuchus spp.) are distributed throughout Mexico and South America. The clade is still in need of revision, but to date, no one has performed extensive molecular analyses on this group. The caimans have the least complete fossil records of all the crocodylians (Brochu, 2003) and most molecular analyses have suffered from incomplete taxon sampling within this group. Nevertheless, the current comparisons using TTR intron 1 place the caimans into the generally accepted phylogeny with high support in all analyses.
Crocodylids comprise the most diverse extant group, and its members occur throughout the tropics and sub tropics on almost every major landmass except for the poles. It contains eleven “true crocodiles” (Crocodylus spp.), the dwarf crocodile (Osteolaemus tetraspis), and the African slender-snouted crocodile (Mecistops cataphractus). This last species has been recently moved from Crocodylus to its own genus (Gatesy and Amato, 2008; McAliley et al., 2006; Willis et al., 2007). Genetic nuclear distances in Table 1 clearly indicate that M. cataphractus has achieved a level of divergence equal to or greater than that seen between Crocodylus and Osteolaemus, which is considered a valid monotypic genus. Furthermore, all analyses place Mecistops as the sister-group to Osteolaemus, which has been suggested by others (Brochu, 1997; Densmore, 1983; Gatesy et al., 2003; White and Densmore, 2000), although most other data sets have not shown the high support that this marker has provided. Molecular and morphological data support the idea that M. cataphractus represents the sole surviving member of an ancient lineage endemic to the African continent (Brochu, 2003).
Finding a marker that can differentiate between the discrepancies in evolutionary divergence times between the crocodylian lineages has been problematic. The true crocodiles (Crocodylus), as well as the Caiman, have split relatively recently (12 and 5 mya respectively), while the alligatorids and gavialids have been recognized by fossil representatives since the late-cretaceous (Brochu, 1999; 2003). Crocodylian systematics is further complicated because the true crocodiles can produce viable offspring through hybridization events and there are instances where the species have sympatric geographic ranges. One would typically infer that these lineages have not been isolated from each other very long. However, there are numerous species which are considered to be related but are indigenous to major continents that have not been connected since the mid-Cretaceous and Jurassic. This would undoubtedly be sufficient to produce reproductive-isolating mechanisms. For this reason, many researchers have proposed a transoceanic hypothesis, which implies that a Crocodylus ancestor arrived in South America approximately 5 million years ago (Brochu et al., 2007). This could explain why Crocodylus forms unresolved clades and has low percent divergence values (0.23%). Even though this seems at first glance to be a highly improbable event, this is not the first time transoceanic events have been proposed to explain evolutionary relationships. Indeed, mammals and other reptiles have also had their share of transoceanic events to explain the evolutionary relationships among the various groups (Carranza and Arnold, 2003; Schrago and Russo, 2003).
The false gharial (Tomistoma schlegelii) has been the most perplexing crocodylian for systematicists. Morphological comparisons of extant and fossil samples consistently place it within the lineage Crocodylidae (Brochu, 1997; Norell, 1989; Vélez-Juarbe et al., 2007), while virtually every molecular analysis classifies it as a gavialid (Gatesy and Amato 2008; Gatesy et al., 2003; Willis et al., 2007). This ambiguity may be a consequence of an incomplete transitional fossil record of this group. Morphological analyses of all known fossils have been robust and have strong support. Again TTR intron 1 corroborates other molecular data by placing Tomistoma schlegelii as the sister taxon to Gavialis gangeticus. The division between these two views seems to suggest that there are still fossils that have yet to be discovered that will coalesce these two hypotheses.
Intron 1 of TTR is best suited for sequencing analysis because it is relatively short across most taxa when compared to the other two introns found in TTR. Additionally, TTR exons 1 and 2 are relatively long and have regions that are highly conserved, which facilitates primer design across diverse species. Although, TTR intron 1 has not been used extensively for molecular analysis, there have been some studies that have shown phylogenetic relationships in a few mammal lineages. Walton et al. (2000) compared the utility of TTR intron 1 to 12S rRNA mitochondrial gene sequences while trying to elucidate the phylogeny of the African mole-rats. This group had previously been examined using 12S rRNA sequences and was found to have several nodes with low bootstrap values. They found that TTR intron 1 increased the resolution of these problematic nodes and produced a more congruent topology. Steiner et al. (2005) used TTR intron 1, interphotoreceptor retinoid binding protein (IRBP), and two mitochondrial genes to elucidate 19 species of didelphid marsupials. Again, TTR intron 1 provided high resolution in several problematic nodes. Additionally, the authors discovered several diagnostic indels among the sequences, which ranged from 5–366 base-pairs that provided unambiguous resolution among several genera. In both papers, the authors compared specimens that were highly divergent between some of the clades. The relatively robust support values in my comparison of crocodylians show that this marker has the potential to be used for other vertebrates that are in need of revision. TTR intron 1 should also be useful for elucidating intergeneric relationships that have minimal divergence. The propensity for this marker to have intergeneric indels would seem ideal for elucidating higher level relationships. As shown previously, these indels appear to be useful as synapomorphic markers for contentious groups (Willis et al., 2007).
Among the vertebrate classes, TTR varies in its tissue-specific expression. TTR is synthesized in the liver and choroid plexus of mammals, marsupials and birds, but it is restricted to the choroid plexus of reptiles. In amphibians and teleosts, on the other hand, TTR is synthesized only in the liver (Achen et al., 1993; Hennebry et al., 2006; Power et al., 2000). These variations in tissue-specific expression imply that there have been different evolutionary pathways for this gene. It certainly appears that stem amniotes mark the first expression of transthyretin in the choroid plexus (Achen et al., 1993).
Comparing the amino acid sequence of TTR in mammals with those of chicken, lizard, frog, and fish reveals the presence of three additional amino acids at the N-terminus in the nonmammalian species: Val-Ser-His in chicken, Crocodylus, and lizards; Gly-Thr-His in frog; and Asp-Lys-His in fish (Power et al., 2000). These changes may be related to functional differences within the protein. Thyroid hormone binding studies have shown that TTR preferentially binds with T3 (triiodothyronine) in birds and T4 (thyroxine) in mammals (Power et al., 2000). This is thought to be due to the changes in the 3′ splice site of intron 1, which has led to the shorter N-terminal amino acid sequence (Aldred et al., 1997). Nevertheless, overall sequence identity among diverse species is relatively high due to the conservative structures of hormone binding sites across almost all taxa.
To date, the TTR gene has been sequenced and submitted to NCBI from only three reptilian species (skink, crocodile, and gecko). There appears to be sufficient sequence conservation between the these samples to design primers for the intron one segment for most other reptilian lineages. However, it is necessary to compare the target product with the conserved TTR flanking exons. The lack of intronic TTR sequences submitted to GenBank (marsupials and mole rats) prevents a direct match in most instances. The sequences available are too divergent to show sufficient similarity to the sequence of interest. This should cease to be a problem as more intron sequences are submitted.
The elucidation of TTR intron one as an effective marker for phylogenetic analysis does not imply that it is to be used as the sole standard in evolutionary relationships. However, the analysis of crocodylian phylogeny with TTR presented here suggests that it offers distinct advantages as a marker. When used in conjunction with other nuclear and mitochondrial markers, as well as fossil and morphological data sets, intron 1 should prove to be useful in the investigation of other closely-related groups.
Acknowledgments
I am indebted to Dr. Llewellyn D. Densmore III for being my Ph.D. mentor and providing me the samples for this work. I thank all the members of the Biotechnology core lab at Texas Tech University for their help and contributions. I especially thank Trisa Crutcher for her cooperation and efficiency in returning sequences in a timely fashion. I thank Dr. Thomas A. Pressley (TAP) for providing me the lab space and support to complete this manuscript. Finally, partial financial support was provided by a grant from the National Institutes of Health (NIH R01 RR10799 to TAP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achen MG, Duan W, Pettersson TM, Harms PJ, Richardson SJ, Lawrence MC, Wettenhall REH, Aldred AR, Schreiber G. Transthyretin gene expression in choroid plexus first evolved in reptiles. Am J Physiol Regulatory Integrative Comp Physiol. 1993;265:982–989. doi: 10.1152/ajpregu.1993.265.5.R982. [DOI] [PubMed] [Google Scholar]
- Aldred AR, Prapunpoj P, Schreiber G. Evolution of shorter more hydrophilic transthyretin N-termini by stepwise conversion of exon 2 into intron 1 sequences (shifting the 3′ splice site of intron 1) European Journal of Biochemistry. 1997;246:401–409. doi: 10.1111/j.1432-1033.1997.t01-1-00401.x. [DOI] [PubMed] [Google Scholar]
- Bayliss P. Survey methods and monitoring within crocodile management programmes. In: Webb GJ, Manolis SC, Whitehead PJ, editors. Wildlife Management: Crocodiles and Alligators. Surrey Beatty and Sons; Sydney, New South Wales, Australia: 1987. [Google Scholar]
- Brochu CA. Morphology, fossils, divergence timing, and the phylogenetic relationships of Gavialis. Syst Biol. 1997;46:479–522. doi: 10.1093/sysbio/46.3.479. [DOI] [PubMed] [Google Scholar]
- Brochu CA. Phylogeny, systematics, and historical biogeography of Alligatoridae. Soc of Vert Paleo Memoir. 1999;6:9–100. [Google Scholar]
- Brochu CA. Phylogenetic approaches toward crocodilian history. Annual Rev Earth Planet Sci. 2003;31:357–397. [Google Scholar]
- Brochu CA, Densmore LD. Crocodile phylogenetics: a review of current progress. In: Grigg G, Seebacher F, Franklin CF, editors. Crocodilian Biology and Evolution. Surrey Beatty and Sons; Sydney, New South Wales, Australia: 2001. [Google Scholar]
- Brochu CA, Nieves-Rivera AM, Vélez-Juarbe J, Daza-Vaca JD, Santos H. Tertiary endemic crocodylians in the West Indies? Geobios. 2007;40:51–59. [Google Scholar]
- Carranza S, Arnold EN. Investigating the origin of transoceanic distributions: mtDNA shows Mabuya lizards (Reptilia: Scincidae) crossed the Atlantic twice. Systematics and Biodiversity. 2003;1:275–282. [Google Scholar]
- Densmore LD. Biochemical and immunological systematics of the order Crocodilia. In: Hecht MK, Wallace B, Prance GH, editors. Evolutionary biology. Vol. 16. Plenum Press; New York: 1983. [Google Scholar]
- Densmore LD, Dessauer HC. Low levels of protein divergence detected between Gavialis and Tomistoma: evidence for crocodilian monophyly. Comp Biochem Physiol B. 1984;77:715–720. [Google Scholar]
- Gatesy J, Amato G. The rapid accumulation of consistent molecular support for intergeneric crocodylian relationships. Molecular Phylogenetics and Evolution. 2008;48:1232–1237. doi: 10.1016/j.ympev.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Amato G, Norell M, DeSalle R, Hayashi C. Combined support for the wholesale taxic atavism in Gavialine crocodylians. Syst Biol. 2003;52:403–422. doi: 10.1080/10635150390197037. [DOI] [PubMed] [Google Scholar]
- Harshman J, Huddleston CJ, Bolback JP, Parsons TJ, Braun MJ. True and false gharials: a nuclear gene phylogeny of crocodylia. Syst Biol. 2003;52:386–402. doi: 10.1080/10635150390197028. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating the human-ape split by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hennebry SC, Wright HM, Likic VA, Richardson SJ. Structural and functional evolution of transthyretin and transthyretin-like proteins. Proteins: Structure, Function and Bioinformatics. 2006;64:1024–1045. doi: 10.1002/prot.21033. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- McAliley LR, Willis RE, Ray DA, White PS, Brochu CA, Densmore LD. Are crocodiles really monophyletic? -Evidence for subdivisions from sequence and morphological data. Mol Phyl Evol. 2006;39:16–32. doi: 10.1016/j.ympev.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Norell MA. The higher level relationships of the extant Crocodylia. J Herpetol. 1989;23:325–335. [Google Scholar]
- Nylander JA. MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Power DM, Elias NP, Richardson SJ, Mendes J, Soares CM, Santos CRA. Evolution of the thyroid hormone-binding protein, transthyretin. General and Comparative Endocrinology. 2000;119:241–245. doi: 10.1006/gcen.2000.7520. [DOI] [PubMed] [Google Scholar]
- Prapunpoj P, Richardson SJ, Schreiber G. Crocodile transthyretin: structure, function, and evolution. Am J Physiol Regulatory Integrative Comp Physiol. 2002;283:R885–R896. doi: 10.1152/ajpregu.00042.2002. [DOI] [PubMed] [Google Scholar]
- Ray DA, Densmore LD. The crocodilian mitochondrial control region: general structure, conserved sequences, and evolutionary implications. J Exp Zool. 2002;294:394–395. doi: 10.1002/jez.10198. [DOI] [PubMed] [Google Scholar]
- Roos J, Aggarwal RK, Janke A. Extended mitogenomic phylogenetic analyses yield new insight into crocodylian evolution and their survival of the Cretaceous-Tertiary boundary. Molecular Phylogenetics and Evolution. 2007;45:663–673. doi: 10.1016/j.ympev.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Schrago CG, Russo CAM. Timing the origin of the New World monkeys. Molecular Biology and Evolution. 2003;20:1620–1625. doi: 10.1093/molbev/msg172. [DOI] [PubMed] [Google Scholar]
- Steiner C, Tilak M, Douzery EJP, Catzeflis FM. New DNA data from a transthyretin nuclear intron suggest an Oligocene to Miocene diversification of living South America opossums (Marsupalia: Didelphidae) Molecular Phylogenetics and Evolution. 2005;35:363–379. doi: 10.1016/j.ympev.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony, Version 4Beta 10. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Vélez-Juarbe J, Brochu CA, Santos H. A gharial from the Oligocene of Puerto Rico: transoceanic dispersal in the history of a non-marine reptile. Proc R Soc B. 2007;274:1245–1254. doi: 10.1098/rspb.2006.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton AH, Nedbal MA, Honeycutt RL. Evidence from the nuclear transthyretin (prealbumin) gene for the phylogeny of African mole-rats (Bathyergidae) Molecular Phylogenetics and Evolution. 2000;16:467–474. doi: 10.1006/mpev.2000.0808. [DOI] [PubMed] [Google Scholar]
- White PS, Densmore LD. A comparison of DNA sequence data analysis methods and their effect on the recovery of crocodylian relationships. In: Grigg G, Seebacher F, Franklin CF, editors. Crocodilian Biology and Evolution. Surrey Beatty and Sons; Sydney, New South Wales, Australia: 2000. [Google Scholar]
- Willis RE, McAliley LR, Neeley ED, Densmore LD. Evidence for placing the false gharial (Tomistoma schlegelii) into the family Gavialidae: inferences from nuclear gene sequences. Molecular Phylogenetics and Evolution. 2007;43:787–794. doi: 10.1016/j.ympev.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Wilgenbusch JC, Warren DL, Swofford DL. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. 2004 doi: 10.1093/bioinformatics/btm388. http://ceb.csit.fsu.edu/awty. [DOI] [PubMed]