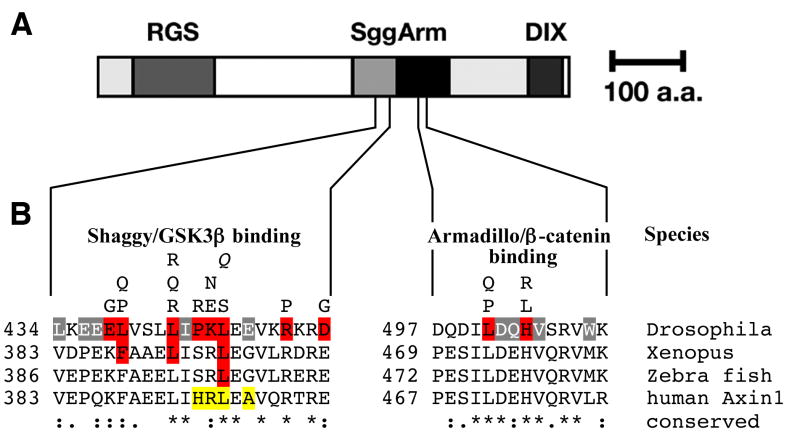
Fig. 1. Identification in vitro of contact residues on Axin critical for binding by Shaggy and Armadillo, respectively.
(A) A schematic Axin structure is shown. Key binding partners and their binding domains are APC (RGS), Shaggy/GSK3β (Sgg), Armadillo/β-catenin (Arm), and the DIX domain required for Axin homodimerization and binding of Dishevelled. In a saturation mutagenesis, we identified the critical residues for Shaggy and Armadillo binding, respectively, using the yeast two hybrid system (Material and methods). (B) An alignment with sequences from the fruit fly (Drosophila melanogaster), frog (Xenopus laevis), zebrafish (Danio rerio) and human is shown. Mutant isolates are shown above the Drosophila sequence and the critical residues are indicated in red. Amino acids highlighted in gray reproducibly disrupted the interaction but depended on the synergy with secondary mutations outside the binding site (Material and methods). L447Q (Q) was not identified as a mutant in the screen but modeled after the zebrafish allele (Heisenberg et al., 2001) and it too disrupts the interaction with Shaggy in yeast. Mutations shown to disrupt the interaction with Xenopus Axin (Hedgepeth et al., 1999) and zebrafish Axin (Heisenberg et al., 2001) are also highlighted in red. Mutations identified in cancers cells but not shown to disrupt the Axin-GSK3β interaction are highlighted in yellow (Salahshor and Woodgett, 2005). The alignment was generated using ClustalW2 and degree of sequence conservation is illustrated (“*”, identity, “:” conserved substitution, “.” semi-conserved substitution).