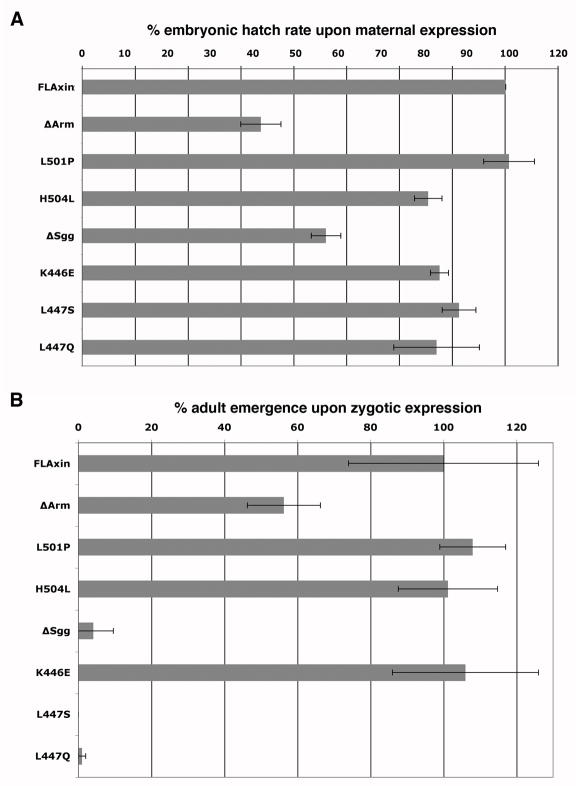
Fig. 7.
Dominant interference with developmental processes by Axin mutant proteins causes death. Mutant Axin proteins expressed at near-physiological levels can disrupt development if they interfere with destruction complex function and result in death (Peterson-Nedry et al., 2008). Conversely, weak Axin alleles do not interfere with the function of endogenous Axin.
(A) The effect of maternally expressed Axin point mutant proteins (tub>Axin*) alongside endogenous wild type Axin is assayed on the ability of embryos to complete development and hatch as larvae and are compared to rates for wild type (wt), full length Axin (FLAxin), as well as the deletion mutants AxinΔArm and AxinΔSgg (Peterson-Nedry et al., 2008). Values are standardize to embryos expressing wild type Axin () and for our reference points (FLAxin, AxinΔArm, AxinΔSgg) were published previously (Peterson-Nedry et al., 2008)). Values are tub>FLAxin, 100% ±1.8, n=554; AxinΔArm, 42.3% ±4.75, n=529; AxinL501P, 101% ±6.0, n=581; AxinH504L 81.9 ±3.3, n= 513; AxinΔSgg 57.8% ±3.5, n=529; AxinK446E, 84.6% ±2.1, n=568; AxinL447S, 89.3% ±4, n=578; AxinL447Q, 83.9% ±10.1, n=569.
(B) Zygotic expression of mutant proteins allows us to assay predominantly late embryonic and post-embryonic development, as the maternal contribution of wild type Axin is depleted. Rates of adult emergence are determined. Again, the mutant proteins are constitutively expressed in the presence of wild type Axin. Here, the point mutants AxinL501P, AxinH504L and AxinK446E do not interfere, while the Shaggy domain point mutants dominantly kill animals. An escaper of Axin L447Q expression is shown in Suppl. Fig. 4. Survival rates are determined by comparing tub>Axin* flies to siblings marked by the slightly detrimental dominant mutation Sp, which results in survival rates greater than 100% if dominant mutations in the control siblings reduce viability; (Materials and methods). Error bars represent standard deviation. Values are compared to FLAxin, AxinΔArm, AxinΔSgg from (Peterson-Nedry et al., 2008) and are derived to the number of surviving non-expressing control sibling (nc): FLAxin, 100% ±26, nc=442; AxinΔArm, AxinL501P, 108% ±9, nc=345; Axin504H, 101.2% ± 13.5, nc=494; 56.3% ±10, nc=417; AxinΔSgg, 4.1% ±5, nc=620; AxinK446E, 106% ±20, nc=362; AxinL447S, 0% ±0, nc=467; AxinL447Q, 1% ±1, nc=366.