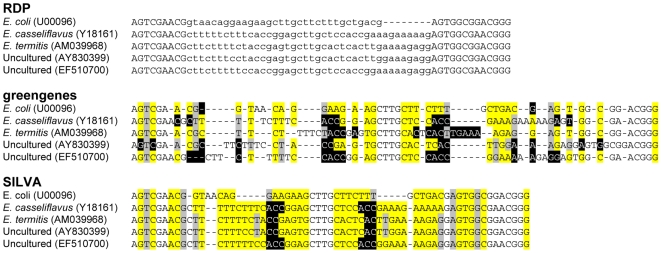
Figure 2. Comparison of alignments generated by the RDP, greengenes, and SILVA databases.
Alignments were taken between positions 60 and 113 of the E. coli 16S rRNA gene sequence for E. coli and four Enteroccocus spp. The alignment generated for these sequences within this region using 8-mers and the Needleman-Wunsch algorithm was identical to that found in the SILVA alignment. The lower-case bases in the RDP alignment indicate unaligned positions. For the greengenes and SILVA alignments, yellow-highlighting represents bases that are predicted to form traditional Watson-Crick base-pairs in the secondary structure, gray-highlighting represents weak base-pairs, black-highlighting represents bases that will not form base-pairs, and a lack of highlighting represents bases that are predicted to be in loop structures.