Table II.
NMR Structural Statistics and Atomic RMS Differences
| Parameters | mCdt1C (420–557) |
|---|---|
| Structure statistics | |
| Completeness bb/sc assign,a % | 99/99 |
| Consensus NOE assign,b % | 64 |
| Total NOE peaks assigned,c (N/Caliphatic/Caromatic) | 3513 (1389/2019/105) |
| NOE constraints | 3295 |
| Short range | 2451 |
| Medium range | 473 |
| Long ranged | 371 |
| No. dihedral angle constraints,e φ/ψ | 94/98 |
| No. hydrogen bond constraints | 86 |
| CYANA target function, Å2 | 2.04 ± 0.28 |
| RMSDs backbone RMSD to mean,f Å | 0.55 ± 0.14 |
| RMSDs heavy atom RMSD to mean,f Å | 0.99 ± 0.11 |
| CNS solve calculation: | |
| RMSDs backbone RMSD to mean,f Å | 0.41 ± 0.07 |
| RMSDs heavy atom RMSD to mean,f Å | 0.92 ± 0.05 |
| Ramachandran plot | |
| Residues in most favored regions, % | 82.7 |
| Residues in additional allowed regions, % | 13.7 |
| Residues in generously allowed regions, % | 2.7 |
| Residues in disallowed regions, % | 0.9g |
For backbone (bb); the assignment yields was calculated by excluding N-terminal 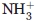 , Pro 15N, and 13C′ shifts of residues preceding Pro residues. For side-chains (sc); excluding side-chain OH, 13C′ and aromatic quaternary 13C shifts, and Lys
, Pro 15N, and 13C′ shifts of residues preceding Pro residues. For side-chains (sc); excluding side-chain OH, 13C′ and aromatic quaternary 13C shifts, and Lys 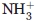 , Arg NH2.
, Arg NH2.
CYANA32 assigned results.
Short-range (|i – j| ≤ 1), medium-range (1 < |i – j| ≤ 5), long-range (|i – j| ≥ 5) assignment with NOE connecting residues i and j.
Calculated by using TALOS.34
RMSDs for well-folded domain, residues 469 to 544.
This result produced with the residues from flexible part.
