Abstract
Fas-associated factor (FAF)-1 is a multidomain protein that was first identified as a member of the Fas death-inducing signaling complex, but later found to be involved in various biological processes. Although the exact mechanisms are not clear, FAF1 seems to play an important role in cancer, asbestos-induced mesotheliomas, and Parkinson's disease. It interacts with polyubiquitinated proteins, Hsp70, and p97/VCP (valosin-containing protein), in addition to the proteins of the Fas-signaling pathway. We have determined the crystal structure of the ubiquitin-associated domain of human FAF1 (hFAF1-UBA) and examined its interaction with ubiquitin and ubiquitin-like proteins using nuclear magnetic resonance. hFAF1-UBA revealed a canonical three-helical bundle that selectively binds to mono- and di-ubiquitin (Lys48-linked), but not to SUMO-1 (small ubiquitin-related modifier 1) or NEDD8 (neural precursor cell expressed, developmentally down-regulated 8). The interaction between hFAF1-UBA and di-ubiquitin involves hydrophobic interaction accompanied by a transition in the di-ubiquitin conformation. These results provide structural insight into the mechanism of polyubiquitin recognition by hFAF1-UBA.
Introduction
Fas-associated factor (FAF)-1 is a 74-kDa novel protein that has multiubiquitin-related domains, which was initially identified as a Fas-associated factor in mice.1 FAF1 is evolutionarily conserved, and in humans it is expressed abundantly in the testes, and slightly less in skeletal muscle and the heart.2 It is also highly expressed in the developing telencephalon with dynamic expression patterns at different embryonic stages, although it progressively becomes confined within limited regions.3
FAF1 was initially identified as a member of the Fas death-inducing signaling complex,1,4,5 but the data accumulated thus far suggest that it is involved in various other biological processes. It acts as a suppressor of NF-κB activity by interfering with nuclear translocation of the RelA subunit of NF-κB.6 Inhibition of NF-κB was also observed in Drosophila7 by caspar, a fly homolog of hFAF1. In Drosophila, caspar was shown to negatively regulate the immune deficiency-mediated immune response by blocking nuclear translocation of NF-κB. FAF1 is reported to be an inhibitor of IκB kinase activation.8 The phosphorylated FAF1 is reported to mediate the ubiquitin-independent, proteasome-dependent degradation of Aurora-A,9 and most recently as a key component of the TNF-α/NF-κB signaling node that has been independently implicated in asbestos-induced oncogenesis through Arf inactivation.10
In addition to being a member of the Fas death-inducing signaling complex, FAF1 reportedly interacts with a number of other proteins. For example, the N-terminal ubiquitin-associated domain of human FAF1 (hFAF1-UBA) is associated with polyubiquitinated proteins in a linkage-specific manner and not to monoubiquitin,11 whereas the UBL1 (ubiquitin-like protein) binds tightly to the N-terminal region of the 70-kDa heat shock protein family, Hsp70/Hsc70, regardless of ATP and heat shock treatment.12 In addition, the C-terminal UBX domain binds the 97kDa valosin-containing protein, a mutiubiquitin chain-targeting factor used in degradation via the ubiquitin-proteasome pathway.11 These findings suggest that FAF1 may serve as a scaffolding protein that regulates protein degradation in the ubiquitin-proteasome pathway.
It is of great interest how one molecule can participate in so many different interactions. As a first step toward understanding this, we have crystallized hFAF1-UBA and analyzed its interactions with ubiquitin and ubiquitin-like proteins using nuclear magnetic resonance (NMR) spectroscopy. The UBA, first identified by sequence database analysis,13 is a commonly occurring ubiquitin-recognition motif that has found in more than 100 human proteins thus far.14–21 It is involved in a wide range of biological activities in signaling pathways including the mediation of cell cycle control, activation of DNA repair mechanisms, and proteasomal degradation.14–16 However, the mechanism of ubiquitin recognition, especially linkage specificity, is still unknown, although some evidence is being accumulated on the recognition of Lys48–linked di-Ub by Mud1-UBA domains17 and hHR23A-UBA.18,41 The three-dimensional solution structure determined by NMR spectroscopy revealed a three-helical bundle.15,18–21
Results and Discussion
The crystal structure of hFAF1-UBA was determined at 1.73 Å resolution, and its interaction with mono- and Lys48-linked di-ubiquitin (denoted as mono-Ub and di-Ub, respectively, hereafter), and SUMO-1 and NEDD8 were examined by using 1H-15N labeled hFAF1-UBA. Furthermore, the binding affinities of the hFAF1-UBA to mono-Ub and di-Ub were measured using NMR and isothermal titration calorimetry (ITC) and the binding surfaces were identified using NMR spectroscopy.
The crystal structure of the hFAF1-UBA
The crystal structure of the UBA domain of hFAF1 (residues 5–47), referred to as hFAF1-UBA (residues 5–47), was solved at 1.73 Å resolution. The structure reveals a canonical UBA-fold consisting of a three-helical bundle (α1, α2, and α3) as shown in Figure 1(A). The α1 helix spans residues Arg7 to Thr18, and α2 spans Ile23 through Asn33, while the α3 consists of Leu37 to Val44. The three helices are tightly packed with a well-defined hydrophobic core that includes Phe14 and Ile20 of α1; Ala26, Leu29, and Leu30 of α2; and Ala40 and Val 44 of α3 [Fig. 1(B)]. The overall structure is quite similar to other UBA domains that have been described thus far, despite the fact that the extent of sequence identities is relatively poor [Fig. 1(C)]. The sequence identity for UBXD8 (PDB entry 2DAM) and p4722 (PDB entry 1V92) are about 38%, whereas others share less than 25%. The root mean square deviations in the backbone range from 0.88 Å for 40 equivalent Cα atoms in UBXD8 to 2.2 Å for 36 equivalent Cα atoms in LATS2 (PDB entry 2COS). These values are comparable with those reported for other UBA structures.
Figure 1.
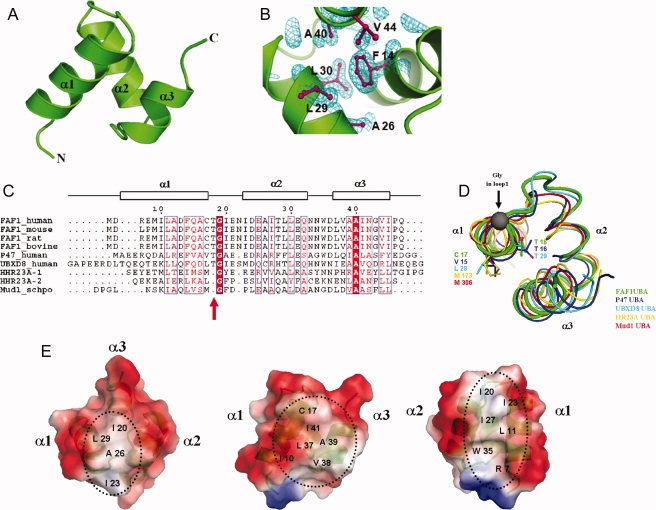
Crystal structure of the hFAF1-UBA. (A) Ribbon representation of hFAF1-UBA. Three helices are labeled as α1, α2, and α3. (B) Electron density (2Fo−Fc map contoured at 1.0σ) of hFAF1-UBA core. The core of the structure is stabilized by hydrophobic residues from all three helices. (C) Sequence alignment of UBA domains. The secondary structure of hFAF1-UBA is indicated by the boxes above the alignment. Residues with more than 70% conservation are boxed. (D) Comparison of the loop connecting α1 and α2. This corresponds to the “Met-Gly-Phe (MGF) motif” found in other UBA domains that has been shown to be involved in ubiquitin binding. hFAF1 is shown in green whereas others are in p47 (1V92) in blue, UBXD8 (2DAM) in cyan, hHR23A (1IFY) in yellow, and Mud1 (1Z96) in red. PDB codes are given in (3E21). (E) Electrostatic potential surface representation of hFAF1-UBA shown at three different orientations.
Comparison with other UBA structures
As expected, hFAF1-UBA showed a compact three-helix bundle. However, this domain shows some significant differences with previously reported UBA domains, the most prominent being the loop connecting α1 and α2, which is formed by Cys17 through Asn22 [Fig. 1(D)]. As seen in the figure, it differs from that is seen in many of the previously reported UBA structures, e.g. hHR23A-UBA15 (PDB entry: 1IFY) and Mud1-UBA17 (PDB entry: 1Z96). In those structures, this loop was noted as the “Met-Gly-Phe” motif, because this region is one of the most conserved stretches in UBA domains.13–21 However, in hFAF1-UBA, it is replaced by four residues, namely Cys-Thr-Gly-Ile, and this is conserved in all reported FAF1 [see Fig. 1(C)]. In addition, Met–Gly–Phe is replaced by Val–Thr–Gly–Ala in p47,22 Ile–Thr–Gly-Ile in UBXD7, and Leu–Thr–Gly–Ile in UBXD8. It is interesting to note the high occurrence of Thr in these non-Met–Gly–Phe UBA domains. The side chain of Thr is situated above Phe, which seems equally conserved. In fact, sequence alignment of UBA domains of a subset of UBX proteins, which were retrieved in a PSI-BLAST search using the shp1 (homologue of p47 in yeast) UBA domain as query sequence, showed high conservation of Thr as well as Phe.23 To examine the significance of this insertion, we made a Thr deletion mutant (referred to as hFAF1-UBA[1–81] (ΔT18) mutant) and this will be discussed later. Nonetheless, the position of the Gly residue remains the same in all cases. In other words, one residue insertion in these UBAs makes extra H-bonds, and therefore a slight extension of the α1-helix [Fig. 1(D)].
The crystal structure reveals a hydrophobic surface between the α1 and α3 helices formed by residues Ile10 from α1, and Leu37 and Ile41 from the α3 helix [Fig. 1(E) middle]. All of the structural studies of UBA and mono-Ub complexes reported so far, e.g. Dsk2-UBA,20 Ede1-UBA,24 and EDD-UBA,25 have identified the hydrophobic patch formed by α1 and α3 of UBA domain as the ubiquitin recognition site, although there may be some variation in the details of the orientation of the two. In all these, the residues of Ub that are involved in UBA binding are Ile44, His68, and Val70. Therefore, it is likely that a similar binding mode occurs between hFAF1-UBA and mono-Ub.
In addition to the canonical hydrophobic patch between the α1 and α3 helices in hFAF1-UBA, there is another hydrophobic patch that is formed between the α1 and α2 helices. Leu11 of α1, Leu23 and Ile27 of α2, and Trp35 form another hydrophobic patch [Fig. 1(E) right]. It is interesting to note that the side chains of the negatively charged residues on α2 are facing the same surface, thus forming a streak of negative surface adjacent to the hydrophobic patch. In fact, the electrostatic surface of hFAF1-UBA is somewhat more acidic than others reported thus far.
Binding of hFAF1-UBA to Ub and ubiqutin-like proteins
To examine whether hFAF1-UBA binds ubiquitin and/or other ubiquitin-like proteins, we have carried out NMR studies using the residues 1–81 of human FAF1, hereafter referred to as hFAF1-UBA [1–81]. In all NMR experiments, hFAF1-UBA [1–81] was used, as opposed to hFAF1-UBA [5–47] used in crystallographic studies, because all biological data reported thus far uses residues 1–81 as the UBA domain. The backbone assignment of hFAF1-UBA [1–81] was performed using conventional triple-resonance experiments and data analysis [Fig. 2(A)]. Chemical shift perturbation (CSP) of 1H-15N HSQC spectra on titration with increasing amount of mono-Ub, SUMO-1, and NEDD8 were measured and the results are shown in Figure 2(B, D, and E, respectively). Lys48-linked di-Ub (referred to as di-Ub hereafter) was also tested [Fig. 2(C)], because Lys48-linked and Lys63-free polyubiquitin chains were mainly identified as a binding partner of hFAF1-UBA when we analyzed the high molecular weight polyubiquitinated proteins interacting with hFAF1 overexpressed in HEK293 cells using nanoLC-ESI-q-TOF tandem MS (see Supporting Information Fig. 1). However, small fraction (<10%) of polyubiquitin chains binding to hFAF1-UBA domain included the diverse-linked polyubiquitin chain containing Lys63- and Lys11-linked ones (data not shown).
Figure 2.
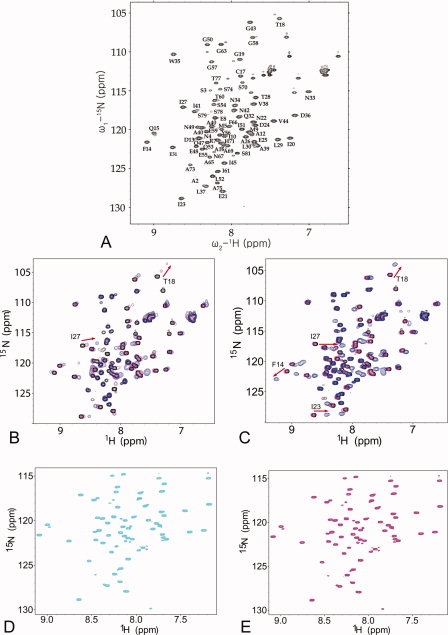
NMR analysis of the interaction of the hFAF1-UBA [1–81] on binding to the ubiquitins and ubiquitin-like proteins. (A) 1H-15N HSQC spectrum of the hFAF1-UBA [1–81] with the backbone assignment. (B–D) Overlays of the 1H-15N HSQC spectra of the hFAF1-UBA [1–81] in the absence and presence of unlabeled binding partner (B) mono-Ub, (C) di-Ub, (D) SUMO-1, and (E) NEDD8. In all the spectra, the peaks representing the free form of the protein are shown in black. In (B), 0.2 mM hFAF1-UBA [1–81] with 0.1 and 1.2 mM mono-Ub were shown in magenta and blue, respectively. In (C), 0.2 mM hFAF1-UBA [1–81] with 0.1 and 0.6 mM di-Ub were shown in magenta and blue, respectively., In (D), 0.5 mM hFAF1-UBA [1–81] with 0.5 mM SUMO-1 was shown in cyan. In (E), 0.5 mM hFAF1-UBA [1–81] with 0.5 mM NEDD8 shown in magenta.
As seen in Figure 2(B and C), certain residues of hFAF1-UBA [1–81] show a CSP to mono- and di-Ub in a dose-dependent manner, whereas there are no significant perturbations on addition of SUMO-1 or NEDD8, thereby indicating that hFAF1-UBA [1–81] specifically binds to mono- and di-Ub but not to SUMO-1 or NEDD8. On titration with increasing amounts of mono-Ub, several resonances shifted to new positions along with the concentration of mono-Ub, indicative of a fast exchange on the chemical shift time scale [Fig. 2(B)]. Slow exchanges are observed between free and bound forms of 15N-hFAF-UBA with NMR signals disappearing at the original position and appearing at a new position [Fig. 2(C)].
The binding affinity of hFAF1-UBA to mono- and di-Ub
We next determined the dissociation constants (Kd) between hFAF1-UBA [1–81] and mono- and di-Ubs using chemical shift titration and ITC. For mono-Ub, a Kd of 1.47 ± 0.10 mM was obtained from the chemical shifts observed for Leu8, Ile13, Ile44, and Val70 of ubiquitin (Fig. 3), whereas the corresponding value was 0.10 ± 0.01 mM for di-Ub following the shifts for Lys6, Thr7, Ile44, and Leu73 of ubiquitin. However, because the later can be erroneous due to the signal broadening and disappearance during the titration, we also measured it using ITC. A Kd value of 3.5 μM was obtained for di-Ub with clear 1:1 binding between the two (Fig. 4), whereas mono-Ub did not show significant binding on ITC. These results show that hFAF1-UBA [1–81] clearly has higher affinity for interaction with di-Ub over mono-Ub. The preference of hFAF1-UBA [1–81] for di-Ub is comparable with Mud1-UBA,17 which showed a Kd of 390 μM for mono-Ub and 3 μM for Lys48-linked di-Ub, or hHR23A-UBA,19 which showed a Kd of 400–500 μM for mono-Ub and 7.7 μM for di-Ub. On the other hand, the UBA domain of p62 binds mono-Ub with higher affinity than di-Ub,21 whereas that of UQ1/PLIC-126 and Mex6727 showed similar affinities with mono- and di-Ub. Thus, it can be suggested that the two ubiquitin domains in the Lys48-linked di-Ub bind to the UBA domain of hFAF1 cooperatively.
Figure 3.
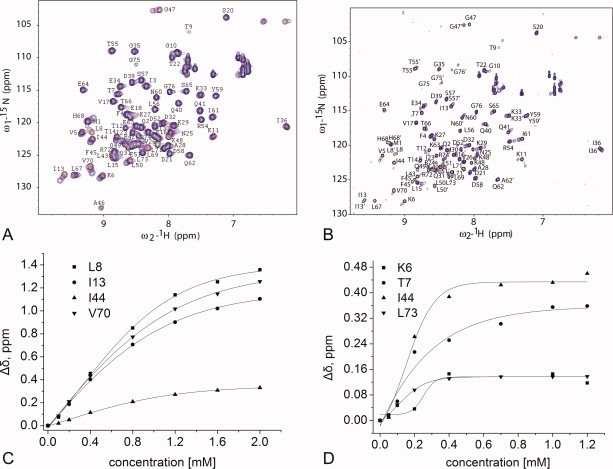
NMR analysis of the interaction of the hFAF1-UBA [1–81] with 15N mono- and di-Ub. Portions of 1H-15N HSQC spectra of 15N-mono-Ub (A) and 15N-di-Ub (B) in the presence of 2 equivalents of unlabeled the hFAF1-UBA [1–81] at 298 K. In (A), 0.2 mM 15N-mono-Ub with 0.1 mM hFAF1-UBA (magenta) and 0.4 mM hFAF1-UBA (blue). In (B), 0.2 mM 15N di-Ub with 0.1 mM hFAF1-UBA (magenta) and 0.4 mM hFAF1-UBA (blue). Plots of the chemical shift changes (Δδ = [(Δδ1H)2 + (Δδ15N/5)2]2) for the selected residues of ubiquitin on the addition of the hFAF1-UBA domain to the (C) mono-Ub and (D) di-Ub. The values of the dissociation constants (Kd) derived from these analyses are 1.47 ± 0.10 mM for mono-Ub and 0.10 ± 0.01 mM for di-Ub, respectively.
Figure 4.
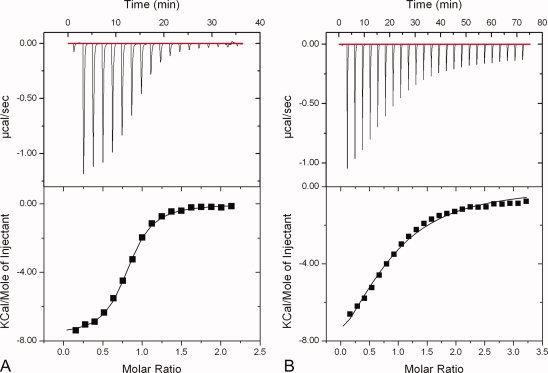
ITC analysis of the interaction between the hFAF1-UBA and di-Ub. (A) Wild-type hFAF1-UBA [1–81] and (B) Thr deletion mutant hFAF1-UBA [1–81] (ΔT18) with di-Ub. The dissociation constants (Kd) are 3.5 and 55 μM, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
However, it is interesting to note that hFAF1-UBA [1–81] (ΔT18) mutant has a Kd value of 55 μM [Fig. 4(B)] and this is more than a 10-fold reduction when compared with wild type. This suggests that the connecting loop between α1 and α2 plays a role in poly-Ub recognition and it is worthwhile mentioning that this Thr residue is conserved in some UBA/UBX proteins, e.g. p47, UBXD7, UBXD8, and FAF1.
NMR mapping of hFAF1-UBA interaction with mono- and di-Ub
We attempted to identify the residues of hFAF1-UBA that are involved in binding using both CSP and cross-saturation. Cross-saturation experiments were carried out since the signal changes in CSP experiments can be attributed not only to direct binding but also as an indirect consequence of structural rearrangement. We also attempted mapping the residues of Ub and di-Ub that are involved in binding using a CSP experiment with labeled mono- and di-Ub.
First, in CSP experiments, the bulk of the strongly perturbed residues (Δσ > 0.1 ppm) was located in the first and second helices and in the loop following the first helix, whereas only a few residues in the third helix were perturbed. In particular, Thr18 and Ile27 of hFAF1-UBA [1–81] show a CSP greater than 0.2 ppm on addition of mono-Ub at 1:6, whereas Phe14, Thr18, Ile 23, and Ile27 of hFAF1-UBA [1–81] show a CSP greater than 0.2 ppm on addition of di-Ub at 1:3 (Supporting Information Fig. S2), suggesting that the similar surface of the hFAF1-UBA is perturbed on binding to mono- and di-Ub.
Second, in cross-saturation experiments, when unlableled mono-Ub was added to 2H, 15N-hFAF1-UBA[1–81], Met9 Asn22, Glu25, Asn34, and Val38 showed signal reductions of more than 10%, although the reduction of peak intensities were relatively weak (∼10% reduction). Resonances showing intensity ratios <0.9 are displayed on the molecular surface, and the major binding surface is present on all three helices as shown in Figure 5(A). Similarly, when cross-saturation measurements were also carried out using unlabelled di-Ub, the result showed more than 60% reduction in peak intensities for the residues at the interface between hFAF1-UBA [1–81] and di-Ub. This includes the residues from all three helices of hFAF1-UBA, namely Met9 and Ile10 (α1); Asp24, Glu25,Ala26, and Glu31 (α2); Ala39, and Ile41 (α3) [see Fig. 5(B)].
Figure 5.
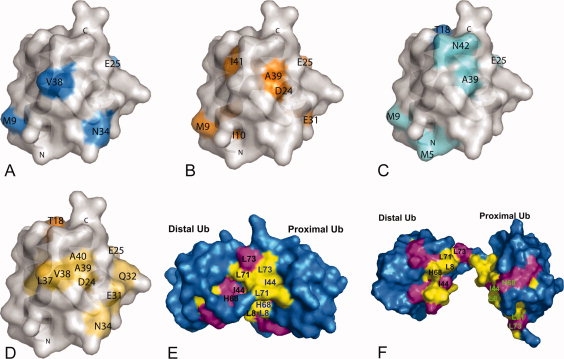
Mapping of the residues involved in binding. (A-B) Mapping of the binding surfaces based on the cross-saturation data of the 1H –15N cross-peaks in 2H, 15N-hFAF1-UBA [1–81] in complex with (A) mono-Ub and (B) di-Ub. Resonances showing intensity ratios <0.9 (for mono-Ub) and <0.4 (for di-Ub) are displayed on the molecular surface of di-Ub in blue and orange, respectively. (C–D) Mapping of the binding surfaces based on the CSP of the 1H –15N cross-peaks in 15N-hFAF1-UBA [1–81] in complex with (C) mono-Ub and (D) di-Ub. Residues are marked in blue (>0.2 ppm) and aquamarine (>0.1 ppm) according to the chemical shift change on hFAF1-UBA [1–81] on the addition of mono-Ub (C) and in orange (>0.1 ppm) and lightorange (>0.05 ppm) according to the chemical shift change on hFAF1-UBA [1–81] on the addition of di-Ub (D). (E–F) Mapping of the molecular surfaces of Lys48-linked di-Ub involved in binding to the hFAF1-UBA [1–81], shown on (E) the closed- and (F) opened conformation of di-Ub part of the complex. Residues that undergo CSP on binding hFAF1-UBA [1–81] are colored in magenta (disappeared peaks) and yellow (Δδ > 0.1 ppm peaks).
When we compare the affected residues on the surface of hFAF1-UBA by CSP and cross-saturation data [Fig. 5(A–D)], they were similar to each other overall. However, there are significant differences that may be due to possible conformational changes of hFAF1-UBA upon binding to mono- or di-Ub, resulting in the additional perturbation of NMR signals. Also, a part of the differences between CSP and cross-saturation results can be attributed to the analysis of cross-saturation data. Because the chemical shifts of the cysteine thiol groups are often near to those of the methyl groups, irradiation of methyl signals may cause the saturation of the thiol hydrogen atoms of free cysteine residues, which may be transferred to the neighboring amide protons. Thus, Gln14, and Thr18, which are located in close proximity to Cys17, were excluded from the mapping.
To map the residues of Ub and di-Ub that are involved in the interaction, we first carried out assignment of di-Ub. We were able to assign the 17 residues from distal and proximal ubiquitin separately in the 1H-15N HSQC spectrum of di-Ub and they were Leu8, Leu43, Phe45, Ala46, Gly47, Lys48, Gln49, Leu50, Glu51, Tyr59, His68, Leu71, Arg72, Leu73, Arg74, Gly75, and Gly76. The other 59 residues were overlapped and thus could not be assigned separately (data will be published elsewhere). The changes in both the 1H-15N HSQC spectra of 15N mono- and di-Ub were analyzed on titration with unlabeled hFAF1-UBA [1–81] [see Fig. 3(A and B), and Supporting Information Fig. S2]. The overall patterns of signal changes of mono- and di-Ub were similar to one another. The observed CSPs in di-Ub occurred in and around the hydrophobic patch consisting of Leu8, Ile44, Gly47, His 68, and Val70. The signals from Leu8, Ile13, Gly47, Gln49, Leu50, Leu67, His68, and Leu71 disappeared, whereas Thr7, Thr14, and Ile44 showed a CSP (Δσ) larger than 3.0 ppm. The presence of significant signal attenuation in di-Ub indicates intermediate or slow exchange on the NMR chemical shift time scale. They are mapped on both the “closed” and “open” structures of Lys48-linked di-Ub as shown Figure 5(E and F, respectively). The closed form is that of the crystal structure28 (PDB entry: 1AAR) and the open form refers to the solution structure of di-Ub complexed with the C-terminal UBA domain of hHR23A18 (PDB entry: 1ZO6). The results show that the most perturbed residues are located at the Ub:Ub interface of the closed state, which indicates that some conformational change of di-Ub affecting its Ub:Ub interface is needed to bind to the UBA domain. Interestingly, most di-Ub residues assigned separately between distal and proximal ubiquitins coincide with the Ub:Ub interface. We suggest that, in the K48-linked di-Ub, the ubiquitin molecules were in close contact with each other, showing the different chemical shifts between distal and proximal ubiquitins, and these were affected by the interaction with hFAF1-UBA, implicating the separation of two ubiquitin molecules from one another.
These results suggest that the UBA binding event is accompanied by a transition in the di-Ub conformation that opens the interface and makes the hydrophobic surfaces of the ubiquitin units available for interaction with UBA, similar to the case between UBA2 of hHR23A and di-Ub.18 The modeled structure of hFAF1-UBA [5–47]:diUb complex based on the CSP and cross-saturation data suggest that the hFAF1-UBA domain is embraced by hydrophobic surfaces on both ubiquitin domains, in a “Ub-UBA-Ub” sandwich-like complex structure (see Supporting Information Fig. S3). Residues from 48 to 81 do not show significant binding and was not counted in this analysis (see Supporting Information Fig. S2). However, the role of the residues 48–81 as a linker between UBA domain and UBLs in hFAF1 should be further studied.
Binding of hFAF1-UBA to poly-Ub in vivo
To confirm the effects of hydrophobic interaction of the UBA domain on the binding with poly-Ub in vivo, we examined the binding of polyubiquitinated proteins with various hFAF1 mutants inside cells. HEK293T cells were transfected with Flag-hFAF1 wild type and various Flag-hFAF1 mutants, e.g. Glu21Ala, Thr28Asn, Val38Asn, Ile41Asn, and Thr18 deletion, and the interacting polyubiquitinated proteins to UBA of Flag-hFAF1 were identified by immunoprecipitation using specific antiflag and western blot analysis using antiubiquitin antibody. The results presented in Figure 6(A) show that the binding of UBA mutants with poly-Ub is significantly less when compared with wild-type UBA, and the Ile41Asn mutant showed the least binding with poly-Ub. It is also worthwhile mentioning that the Thr18 deletion mutant shows a significant reduction in poly-Ub binding; as much as that of Ile41Asn mutant [Fig. 6(B)]. This is in agreement with the earlier ITC data that showed more than a 10-fold reduction in binding affinity compared with wild type (Fig. 4).
Figure 6.
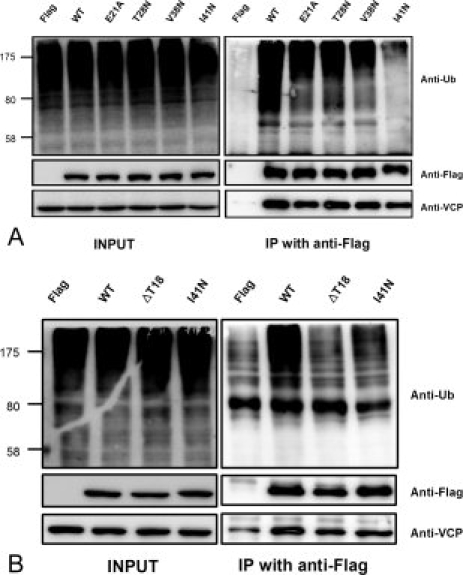
Various mutants of hFAF1-UBA binds to polyubiquitin chain in a different way in vivo HEK293T cells transfected with various mutant Flag-hFAF1s; (A) Flag-hFAF1 wild type, Glu21Ala, Thr28Asn, Val38Asn, and Ile41Asn. (B) Flag-hFAF1 wild type, ΔThr18 and Ile41Asn, were immunoprecipitated with antiFlag antibody and binding polyubiqutinated proteins were detected with antiubiquitin antibody.
Conclusion
The hFAF1-UBA domain adopts a canonical three-helical bundle that is similar to the previously reported UBA domain. It specifically recognizes mono- and di-Ub, and not SUMO-1 or NEDD8. The Kd values between the hFAF1-UBA [1–81] and mono-Ub and di-Ub are 1.47 ± 0.10 mM and 3.5 μM, respectively, and they are comparable with those observed for other UBA domains. The analysis of the interaction between the UBA and Lys48-linked di-Ub suggests a “sandwich-like” structure, with UBA in the middle. Site-directed mutagenesis studies provided supporting evidence of the importance of the hydrophobic residues of the UBA domain for poly-Ub binding. These results provide structural insight into the mechanism of poly-Ub recognition by the hFAF1-UBA domain.
Materials and Methods
Cloning, protein expression, purification, and sample preparation
For crystal structure, the UBA domain of hFAF1 (residues 5–47), referred to as hFAF1-UBA [5–47], was cloned into a pGEX-4T-1 vector (GE Helathcare) and expressed in Escherichia coli BL21 (DE3) in rich Luria-Bertani medium as a fusion with N-terminal GST (glutathione S-transferase) tag. GST fusion protein was purified by affinity chromatography using a GST affinity column (GE Helathcare), and the tag was removed by cleavage with thrombin. The cleaved protein was further purified using Superdex 75s column (GE Healthcare) and concentrated to 25 mg/mL using an Amicon Ultra-15 concentrator (Millipore).
For NMR analysis, the residues 1–81 of human FAF1, referred to as hFAF1-UBA [1–81], was cloned into a pGEX-4T-1 (GE Healthcare) and expressed in Escherichia coli BL21 (DE3). Cells were grown in 15NH4Cl-M9 or M9 medium supplemented with 15NH4Cl and 2H6-glucose in 99.9% D2O at 37°C to OD600∼0.6, and induced using 300 mM IPTG (isopropyl-β-D-1-thiogalactopyrano-side) and allowed to grow overnight with agitation at 18°C. N-terminal GST-fused hFAF1-UBA [1–81] was purified using the same procedure as the hFAF1-UBA[5–47] domain. The final buffer condition for NMR study was 20 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.0, 100 mM NaCl, and 1 mM DTT.
Mouse E1, human Ubc7, mono-Ub, and NEDD8 were cloned, expressed, and purified as described earlier.29,30 A K48-linked di-Ub was prepared by Ubc7-mediated in vitro ubiquitination in the absence of E3, and the preparation was then subjected to Superdex 75s column. The fractions that contained mainly di-Ub were purified further on a Mono S column (GE Healthcare) with a linear gradient from 0 to 1M NaCl at pH 4.5.31
Crystallization, data collection, and structure determination of hFAF1-UBA domain
Crystals were obtained by mixing a ∼25 mg/mL protein in 50 mM HEPES, pH 7.5, 150 mM NaCl and 1 mM DTT, with a reservoir solution of 25% w/v PEG3350 and 200 mM ammonium nitrate. They were stabilized in 25% ethylene glycol and flash frozen before data collection. Diffraction data from a single crystal were collected using synchrotron radiation (Pohang Light Source 4A beamline) at 100 K using an ADSC Quantum 210 CCD area detector system (ADSC, USA). The raw data were processed and scaled using the program suite HKL200032 and the statistics on data collection are given in Table I.
Table I.
Data Collection and Refinement Statistics Values in Parentheses are for the Outer Most Resolution Shell (1.73–1.70 Å)
| hFAF-1 UBA | |||||
|---|---|---|---|---|---|
| Wavelength (Å) | 1.0000 Å. (PAL, 4A MXW) | ||||
| Resolution range (Å) | 50–1.73 | ||||
| Space group | P212121 | ||||
| Unit cell parameters | a = 31.515 Å | ||||
| b = 34.757 Å | |||||
| c = 36.129 Å | |||||
| α = β = γ = 90.0° | |||||
| Total/unique reflections | 230247 (4463) | ||||
| Completeness (last shell) (%) | 98.2 (96.8) | ||||
| Mean I/σ(I) (last shell) (%) | 48.5 (6.7) | ||||
| Rmergea (last shell) (%) | 6.1 (15.5) | ||||
| Refinement | RMSD from Ideality | Ramachandran Analysis | |||
| Resolution range (Å) | 20–1.73 | Bond lengths (Å) | 0.0040 | Favored (%) | 91.7 |
| Allowed (%) | 8.3 | ||||
| R value/R free | 21.8/22.2 | Bond angles (°) | 1.0295 | Generally allowed (%) | 0 |
| No. protein atoms | 342 | Disallowed (%) | 0 | ||
| No. water molecules | 32 | ||||
Rmerge = ∑h∑i|I(h,i) − 〈I(h)〉| / ∑h∑iI(h,i), where I(h,i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the mean value of I(h,i) for all i measurements.
The structure of the hFAF1-UBA was determined by molecular replacement with CNS, using the UBA domain of UBX-domain containing protein 8 (UBXD8), protein ETEA (PDB entry 2DAM). The initial model obtained was adjusted using the program COOT33 and refinement was carried out using CNS.34 The final model contains all residues except the three residues at the C-terminus and 32 water molecules. The statistics on the final refinement are given in Table I.
NMR spectroscopy
CSPs were monitored in a series of 1H-15N HSQC spectra as unlabeled mono-Ub and di-Ub, SUMO-1, and NEDD8 were added to 15N-hFAF1-UBA [1–81]. Protein samples were prepared in 20 mM HEPES buffer, pH 7.0, 100 mM NaCl, 1 mM DTT, and 10% (w/v) D2O. The experiments were performed by titrating 0.2-0.5 mM 15N-labeled protein with concentrated stocks of the appropriate unlabeled binding partners. We used general triple-resonance experiments for the backbone assignment of hFAF1-UBA [1–81].35 Amide CSPs were calculated as Δδ = [(Δδ1H)2+(Δδ15N/5)2]2, where Δδ1H and Δδ15N are the observed chemical shift changes for 1H and 15N, respectively.40
The equilibrium dissociation constants were measured from the curve fitting of CSPs observed between labeled protein and ligand as described by Varadan et al.18 The cross-saturation experiments were performed using the pulse scheme shown by Takahashi et al.,36 on a sample containing 0.4 mM2H, 15N-labeled hFAF-UBA [1–81] in complex with 0.2 mM unlabeled mono- or Lys48-linked di-Ub, and 90% D2O in the NMR buffer. Saturation of the aliphatic protons of the mono-or di-Ub was done using the WURST-2 decoupling scheme.37 The saturation frequency was set at 0.9 ppm. The recycle delay and saturation time were set to 3.2 and 1.2 s, respectively. All NMR experiments were carried out at 25°C using a Bruker Avance500 spectrometer equipped with a cryogenic probe and Bruker Avance800 spectrometer in Korea Basic Science Institute.
Isothermal titration calorimetry
ITC experiments were performed using a ITC200 instrument (MicroCal., Inc., USA). Purified proteins were dialyzed into 20 mM HEPES buffer, pH 7.5, 100 mM NaCl. Titrations were performed by injecting of hFAF1-UBA [1–81] wild type (2 mM) and hFAF1-UBA [1–81] (ΔT18) mutant (2 mM) into the sample cell containing di-Ub (0.1 mM) at 25°C. The binding stoichiometry and binding constants were determined by fitting the data to a one-site binding model. The ITC data were fit using Origin 7.0 (MicroCal., Inc., USA).
Mass spectrometric analysis of polyubiquitin
The silver-stained gel bands were destained and digested with trypsin and peptides extracted as described previously.38 The extracted solutions were evaporated to dryness in Speedvac for MS analysis. Formic acid was then added to the final solution to a final concentration of 0.1% acid in solvent to facilitate electrospray. Peptide analyses were performed by nano flow reversed-phased LC-ESI MS/MS spectrometer (Q-tof UltimaTM global, Waters Co., UK). Peptides were separated using a C18 reversed-phase 75 μm i. d. × 150 mm analytical column (3 μm particle size, AtlantisTM dC18, Waters) with an integrated electrospray ionization SilicaTipTM (±10 μm i.d., New Objective, USA). Five microliters of peptide mixtures were dissolved in buffer C (water/ACN/formic acid; 95:5:0.2, v/v), injected on a column and eluted with a linear gradient of 5–80% buffer B (ACN/water/formic acid; 95:5:0.2, v/v) over 120 min. Samples were desalted online before separation using trap column (5-μm particle size, NanoEaseTM dC18, Waters) cartridge. Initially, the flow rate was set to 250 nL/min by split/splitless inlet and the capillary voltage (3.0 keV) was applied to the LC mobile phase before spray. MS parameters for efficient data-dependent acquisition were as follows: intensity (>10), and three to four components to be switched from MS to MS/MS analysis. The raw data files obtained from the mass spectrometer were converted to.pkl files using ProteinLynx 2.1 (Waters Co., UK). The.pkl files were searched against a SWISS-PROT database using Mascot (global search engine). Precusor ion mass corrections and a fragment ion mass tolerance of 0.2 Da were used, considered to two missed cleavages, enzyme limited to trypsin, toxonomy limited to homo sapiens, fixed modifications unconsidered and variable modifications considered were acetylation, deamidation, ubiqutination (GlyGly), methylation, pyro-glu (N-term E, Q), oxidation, phosphorylation, and cysteine propionamide.
Transient transfection and immunoblot analysis
Human embryonic kidney epithelial cells (HEK293T) were grown and maintained at 37°C and 5% CO2 in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Monoclonal antiFlag antibody M2 was purchased from Sigma (USA), and monoclonal antiubiquitin antibody from Chemicon (Temecula, CA). HEK293T cells (1.5 × 106) were seeded in 100-mm dishes and transiently transfected the next day with 6 to 12 μg of expression plasmids by the calcium phosphate method. The medium was changed with fresh 10% fetal bovine serum-containing Dulbecco's modified Eagle's medium for 6 h and cultured for an additional 18 h before harvesting or further treatment. Mutants of hFAF1 in UBA domain (Glu21Ala, Thr28Asn, Val38Asn, Ile41Asn, and Thr18 deletion) were constructed by site-directed mutagenesis of using the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA).
For western analysis, cells were lysed with lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, and 5 mM DTT) containing 10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 μg/mL pepstatin, and 100 μg/mL phenylmethylsulfonyl fluoride, 10 mM NaF, 10 mM Na3VO4 for 30 min on ice. The protein concentrations of cell lysates were measured by Bradford assay (Bio-Rad Laboratories, Hercules, CA). Proteins (30 μg) were separated by SDS-PAGE under reducing conditions, transferred to PVDF membrane, and probed with monoclonal antibody to Flag (Sigma, USA), polyclonal antibody to FAF full or [1–81], and monoclonal antiubiquitin antibody from Chemicon (Temecula, CA). The protein–antibody complexes were visualized with horseradish peroxidase–conjugated secondary antibody (Cell Signaling Technology, USA) for 1 h at a 1:2000 dilution. The blots were incubated for 3 min in ECL plus kit (GE Healthcare, UK) and exposed to LAS3000 (Fuji Photo Film Co., Japan).
Electronic supplementary material is available for mass spectrometric analysis of polyubiquitin and CSP and cross-saturation data from binding of the hFAF1-UBA[1–81] with mono- and di-Ub as well as a docked model of the hFAF1-UBA: di-Ub complex.
Protein data bank accession codes
The atomic coordinates and structure factors of the crystal structure have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Bruinswick, NJ (http://www.rcsb.org/), with accession number 3E21.
Acknowledgments
The authors thank the staffs at the 4A beamline of PAL, for assistance in data collection.
Glossary
Abbreviations:
- hFAF1
human Fas-associated factor 1
- Ub
Ubiquitin
- di-Ub
Lys48-linked di-ubiquitin
- RMSD
root mean square deviation
- CSP
chemical shift perturbation.
References
- 1.Chu K, Niu X, Williams LT. Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc Natl Acad Sci USA. 1995;92:11894–11898. doi: 10.1073/pnas.92.25.11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryu SW, Chae SK, Lee KJ, Kim E. Identification and characterization of human Fas associated factor 1, hFAF1. Biochem Biophys Res Commun. 1999;262:388–394. doi: 10.1006/bbrc.1999.1217. [DOI] [PubMed] [Google Scholar]
- 3.De Zio D, Ferraro E, D'amelio M, Simoni V, Bordi M, Soroldoni D, Berghella L, Meyer BI, Cecconi F. Faf1 is expressed during neurodevelopment and is involved in Apaf1-dependent caspase-3 activation in proneural cells. Cell Mol Life Sci. 2008;65:1780–1790. doi: 10.1007/s00018-008-8075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu SW, Lee SJ, Park MY, Jun JI, Jung YK, Kim E. Fas-associated factor 1, FAF1, is a member of Fas death-inducing signaling complex. J Biol Chem. 2003;278:24003–24010. doi: 10.1074/jbc.M302200200. [DOI] [PubMed] [Google Scholar]
- 5.Park MY, Ryu SW, Kim KD, Lim JS, Lee ZW, Kim E. Fas-associated factor-1 mediates chemotherapeutic-induced apoptosis via death effector filament formation. Int J Cancer. 2005;115:412–418. doi: 10.1002/ijc.20857. [DOI] [PubMed] [Google Scholar]
- 6.Park MY, Jang HD, Lee SY, Lee KJ, Kim E. Fas-associated Factor-1 inhibits nuclear factor-kappaB (NF-kappaB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-kappaB. J Biol Chem. 2004;279:2544–2549. doi: 10.1074/jbc.M304565200. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci USA. 2006;103:16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park MY, Moon JH, Lee KS, Choi HI, Chung J, Hong HJ, Kim E. FAF1 suppresses IkB kinase (IKK) activation by disrupting the IKK complex assembly. J Biol Chem. 2007;282:27572–27577. doi: 10.1074/jbc.C700106200. [DOI] [PubMed] [Google Scholar]
- 9.Jang MS, Sul JW, Choi BJ, Lee SJ, Suh JH, Kim NS, Kim WH, Lim DS, Lee CW, Kim E. Negative feedback regulation of Aurora-A via phosphorylation of Fas-associated factor-1. J Biol Chem. 2008;283:32343–32351. doi: 10.1074/jbc.M804199200. [DOI] [PubMed] [Google Scholar]
- 10.Altomare DA, Menges CW, Pei J, Zhang L, Skele-Stump KL, Carbone M, Kane AB, Testa JR. Activated TNF-alpha/NF-kappaB signaling via down-regulation of Fas-associated factor 1 in asbestos-induced mesotheliomas from Arf knockout mice. Proc Natl Acad Sci USA. 2009;106:3420–3425. doi: 10.1073/pnas.0808816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song EJ, Yim SH, Kim E, Kim NS, Lee KJ. Human Fas assocated factor 1, interacting with ubiquitinated proteins and valosin-containing protein, is involved in ubiquitin-proteasome pathway. Mol Cell Biol. 2005;25:2511–2524. doi: 10.1128/MCB.25.6.2511-2524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HJ, Song EJ, Lee YS, Kim E, Lee KJ. Human Fas-associated factor 1 interacts with heat shock protein 70 and negatively regulates chaperone activity. J Biol Chem. 2005;280:8125–8133. doi: 10.1074/jbc.M406297200. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann K, Bucher P. The UBA domains: a sequence motif present in multiple enzyme classes of the ubiqutination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 14.Wilkinson CR, Seeger M, Hatmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:9732–9742. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 15.Mueller TD, Kamionka M, Feigon J. Specificity of the Interaction between Ubiquitin associated domains and Ubiquitin. J Biol Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 16.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 17.Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 20.Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure. 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- 22.Yuan X, Simpson P, Mckeown C, Kondo H, Uchiyama K, Wallis R, Dreveny I, Keetch C, Zhang X, Robinson C, Freemont P, Matthews S. Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97. EMBO J. 2004;23:1463–1473. doi: 10.1038/sj.emboj.7600152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuberth C, Richly H, Rumpf S, Buchbeger A. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 2004;5:818–824. doi: 10.1038/sj.embor.7400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson KA, Hicke L, Radhakrishnan I. Structural basis for monoubiquitin recognition by the Ede1 UBA domain. J Mol Biol. 2006;358:713–724. doi: 10.1016/j.jmb.2006.02.059. [DOI] [PubMed] [Google Scholar]
- 25.Kozlov G, Nguyen L, Lin T, De Crescenzo G, Park M, Gehring K. Structural basis of ubiquitin recognition by the ubiquitin-associated (UBA) domain of the ubiquitin ligase EDD. J Biol Chem. 2007;282:35787–35795. doi: 10.1074/jbc.M705655200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2009;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobeika M, Brockmann C, Iglesias N, Gwizdek C, Neuhaus D, Stutz F, Stewart M, Divita G, Dargemont C. Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67. Mol Biol Cell. 2007;18:2561–2568. doi: 10.1091/mbc.E07-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 29.Ryu KS, Choi YS, Ko J, Kim SO, Kim HJ, Cheong HK, Jeon YH, Choi BS, Cheong C. Direct characterization of E2-dependent target specificity and processivity using an artificial p27-linker-E2 ubiquitination system. BMB Rep. 2008;41:852–857. doi: 10.5483/bmbrep.2008.41.12.852. [DOI] [PubMed] [Google Scholar]
- 30.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 31.Pickart CM, Haldeman MT, Kasperek EM, Chen Z. Iodination of tyrosine 59 of ubiquitin selectively blocks ubiquitin's acceptor activity in diubiquitin synthesis catalyzed by E2(25K) J Biol Chem. 1992;267:14418–14423. [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D. 2004;60:2126–2123. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstlever RW, Jian JS, Kuszewki J, Nigles M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Shimada I, Ueda T, Matsumoto M, Sakakura M, Osawa M. Cross-saturation and transferred cross-saturation experiments. Prog NMR Spectrosc. 2009;54:123–140. [Google Scholar]
- 36.Takahashi H, Nakanishi T, Kami K, Arata Y, Shimada I. A novel NMR method for determining the interfaces of large protein–protein complexes. Nature. 2000;7:220–223. doi: 10.1038/73331. [DOI] [PubMed] [Google Scholar]
- 37.Kupce E, Wagner GW. Homonuclear decoupling in protein spectra. J Magn Res B. 1995;109:329–333. [Google Scholar]
- 38.Seo J, Jeong J, Kim YM, Hwang N, Paek E, Lee KJ. Strategy for comprehensive identification of post-translational modifications in cellular proteins, including low abundant modifications: application to glyceraldehyde-3-phosphate dehydrogenase. J Proteome Res. 2008;7:587–602. doi: 10.1021/pr700657y. [DOI] [PubMed] [Google Scholar]
- 39.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 40.Kim KA, Song JS, Jee J, Sheen MR, Lee C, Lee TG, Ro S, Cho JM, Lee W, Yamazaki T, Jeon YH, Cheong C. Structure of human PRL-3, the phosphatase associated with cancer metastasis. FEBS Lett. 2004;565:181–187. doi: 10.1016/j.febslet.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 41.Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J Biol Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]


