Figure 1.
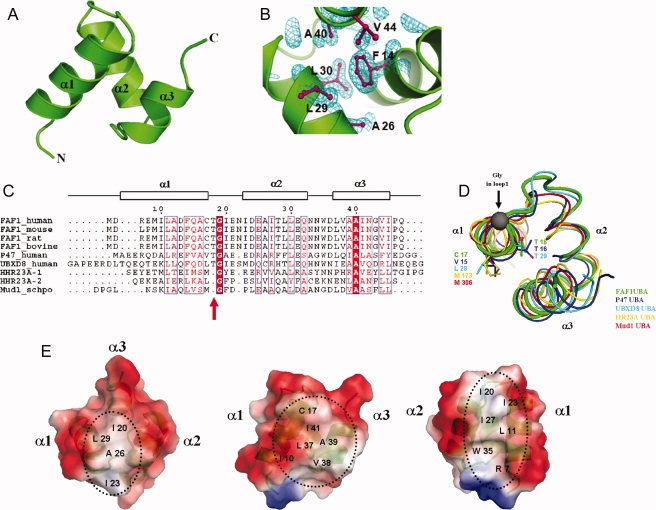
Crystal structure of the hFAF1-UBA. (A) Ribbon representation of hFAF1-UBA. Three helices are labeled as α1, α2, and α3. (B) Electron density (2Fo−Fc map contoured at 1.0σ) of hFAF1-UBA core. The core of the structure is stabilized by hydrophobic residues from all three helices. (C) Sequence alignment of UBA domains. The secondary structure of hFAF1-UBA is indicated by the boxes above the alignment. Residues with more than 70% conservation are boxed. (D) Comparison of the loop connecting α1 and α2. This corresponds to the “Met-Gly-Phe (MGF) motif” found in other UBA domains that has been shown to be involved in ubiquitin binding. hFAF1 is shown in green whereas others are in p47 (1V92) in blue, UBXD8 (2DAM) in cyan, hHR23A (1IFY) in yellow, and Mud1 (1Z96) in red. PDB codes are given in (3E21). (E) Electrostatic potential surface representation of hFAF1-UBA shown at three different orientations.
