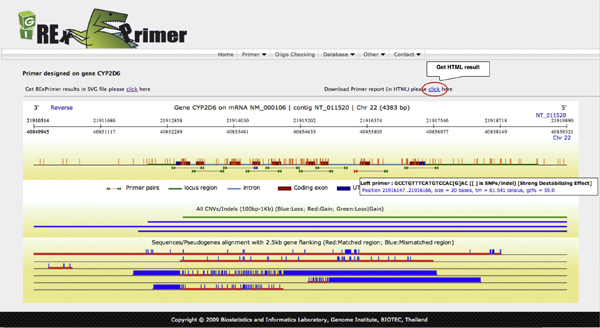
Figure 2.
The SVG graphical output displaying resulting primer pairs with other genomic features. The designed primer pairs are represented by double-headed arrows along with gene structure (intron/exon) and other genomic features (pseudogenes, indels, CNVs, etc.). Pseudogenes and CNVs/indels are shown as multiple sequence alignments with the target gene. SNP-in-Primers are demonstrated with different colors according to the degree of destabilizing effects introduced by such SNPs. Green color presents primer without SNPs inside. Blue color represents primer with SNP in any position but not within 7 bp of the 3'end while red color specifies primer with SNP found within 7 bp of the 3'end. Each designed primer pair can be linked-out to a redesign module.