Figure 1.
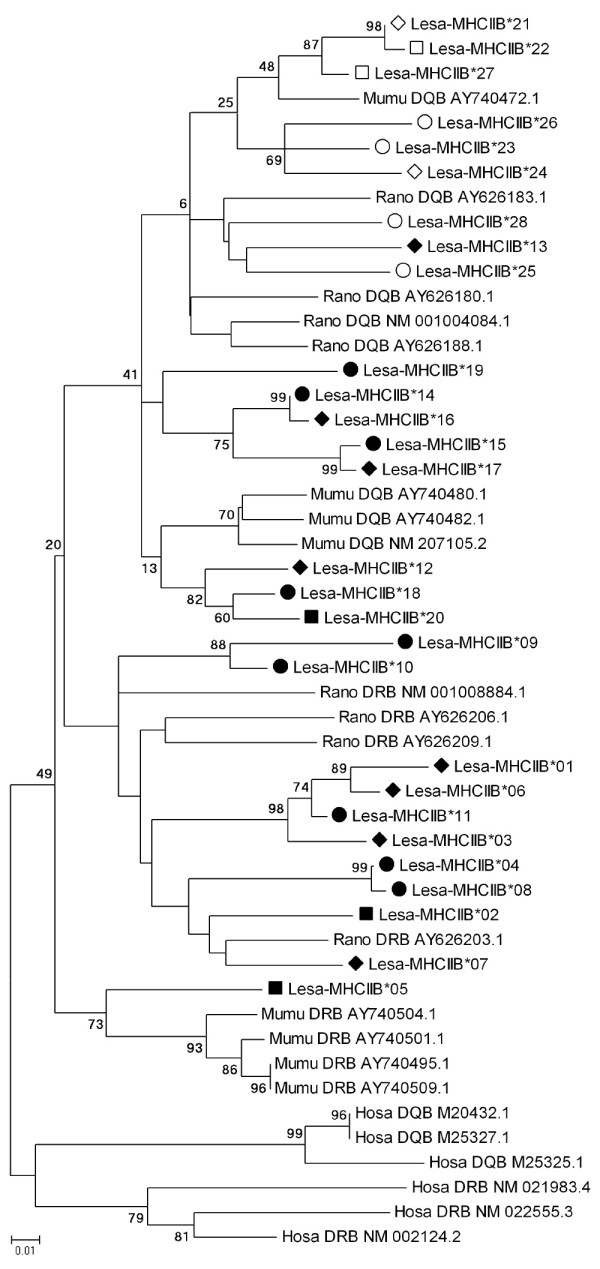
Neighbour-joining tree (bootstrap consensus) of MHC class IIB alleles. Phylogenetic tree based on NJ algorithms calculated at the nucleotide level; including Leopoldamys sabanus (Lesa) and non-Lesa alleles (Hosa - Homo sapiens, Rano - Rattus norvegicus, Mumu - Mus musculus). Alleles occurred in both subpopulations (◆), only in Poring (■) or only in Monggis (●). Open symbols indicate alleles with a deletion of two amino acids. For the non-basal nodes, only bootstrap values above 50 are shown.
