Figure 1.
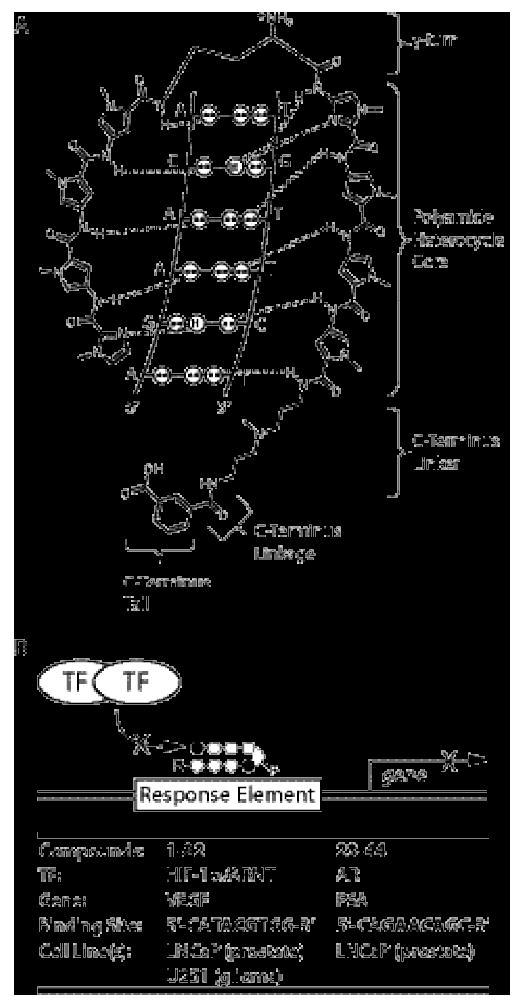
Project overview. A) Schematic illustration of polyamide-DNA recognition through formation of hydrogen bonds with the floor of the DNA minor groove. The γ-turn, polyamide heterocycle core, C-terminus linker, C-terminus linkage, and C-terminus tail of the polyamide are indicated by brackets and labeled for clarity. B) Schematic illustration of the mechanism by which polyamides affect gene expression. Sequence-specific binding of a polyamide core to a gene Response Element (RE) blocks Transcription Factor (TF) binding to the RE, thus blocking up-regulation of gene product expression under inducing conditions. Relative mRNA expression levels under inducing conditions were employed as a biological readout of polyamide cell uptake nuclear localization. Displayed in tabular form are the relevant transcription factors, gene products, DNA RE binding sequence, and cell lines for each set of match and mismatch polyamides.
