Figure 5.
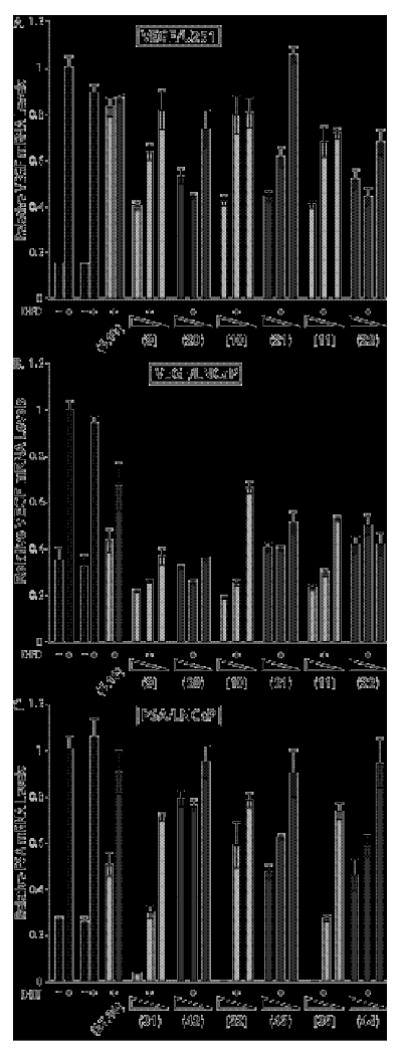
Gene-regulation effects measured by quantitative real-time RT-PCR for the oxime-linked polyamides 9-11, 20-23, 31-33 and 42-44, and the hydroxylamine tail control compounds 5, 16, 27, and 38. Induced and uniduced control conditions are indicated by black bars, match core compounds by light grey bars, and mismatch core compounds by dark grey bars. Uninduced and induced control compounds without DMSO are on the left, and with 0.1% DMSO on the right. Errors shown are the fractional standard deviation. A) Effect on DFO-induced expression of VEGF by HRE-targeted match and mismatch polyamide cores in U251 cells at 1, 0.2, and 0.02 μM; B) Effect on DFO-induced expression of VEGF by HRE-targeted match and mismatch polyamide cores in LNCaP cells at 10, 2 and 1 μM; C) Effect on DHT-induced expression of PSA mRNA by ARE-targeted match and mismatch polyamide cores in LNCaP cells at 5, 1 and 0.1 μM.
