SUMMARY
HOXB13 is a member of the homeodomain family of sequence-specific transcription factors and together with the androgen receptor (AR) plays a critical role in the normal development of the prostate gland. We demonstrate here that in prostate cancer cells HOXB13 is a key determinant of the response to androgens. Specifically, it was determined that HOXB13 interacts with the DNA binding domain of AR and inhibits the transcription of genes that contain an androgen response element (ARE). In contrast, the AR:HOXB13 complex confers androgen responsiveness to promoters that contain a specific HOXB13-response element. Further, HOXB13 and AR synergize to enhance the transcription of genes that contain a HOX element juxtaposed to an ARE. The profound effects of HOXB13 knockdown on androgen regulated proliferation, migration and lipogenesis, in prostate cancer cells highlight the importance of the observed changes in gene expression.
INTRODUCTION
The androgen receptor (AR), a member of the nuclear receptor (NR) superfamily of ligand-regulated transcription factors, is a key regulator of male reproductive function and plays a pivotal role in the development and maintenance of the prostate gland. In addition, AR is also linked to the progression of prostate cancer, a disease that affects one in six men during their lifetimes (Jemal et al., 2006). Upon binding either testosterone (T) or dihydrotestosterone (DHT), the receptor undergoes an activating conformational change that leads to the dissociation of an inhibitory heat shock protein complex, followed by translocation of the receptor to the nucleus and its subsequent interaction with the regulatory regions of target genes. Promoter context and the relative and absolute level of coactivators and corepressors are key determinants of the resulting transcriptional activity of the DNA-bound receptor.
A significant number of coregulatory proteins have been identified that participate in AR signaling by remodeling chromatin structure and facilitating the recruitment of RNA polymerase II to target gene promoters. For instance, ligand-bound AR has been shown to recruit the p160 family of transcriptional activators (SRC1, GRIP1, ACTR), proteins that possess histone acetyltransferase (HAT) activity, to the promoter of the androgen-responsive PSA gene (Bevan et al., 1999; Ma et al., 1999; Wang et al., 2005). Other coregulatory proteins possessing enzymatic activity have been identified that interact with AR or its associated p160 proteins including CARM1 (methyltransferase), LSD1 (lysine specific demethylase), and CBP/p300 (HAT activity) (Koh et al., 2002; Metzger et al., 2005; Wang et al., 2005). AR can also recruit TRAP220, a subunit of the mediator complex, to target genes providing a direct point of contact with components of the basal transcription machinery (Wang et al., 2002). More recently, the homeobox protein, HOXB13, has been shown to interact with and suppress AR transcriptional activity and androgen-mediated prostate cancer cell growth when overexpressed in cells (Jung et al., 2004).
Homeodomain containing proteins (HOX) are the second largest class of sequence-specific transcription factors (Hueber and Lohmann, 2008). They contain a highly conserved DNA-binding domain, the homeobox, which consists of approximately 60 amino acids. Recent studies, in both Drosophila and mice, reveal a core DNA binding sequence rich in adenosine and thymidine nucleotides (TAAT) although individual homeodomains have distinct sequence binding preferences (Berger et al., 2008; Noyes et al., 2008). In humans, there are 39 HOX genes arrayed in chromosomal clusters designated HOXA, HOXB, HOXC, and HOXD. The genes in each cluster are arranged into 13 paralog groups and their position in the genome (3’ to 5’) determines their expression along the anterior-posterior (AP) axis (from posterior to anterior) (Hueber and Lohmann, 2008). Indeed, one of the primary functions of the HOX family of transcription factors during development is to specify the identity of body segments along the AP axis.
HOXB13 is expressed predominantly in the tailbud and posterior domains of the spinal cord, digestive tract and urogential sinus (Zeltser et al., 1996). Not surprisingly therefore, HOXB13 was found to play an essential role in prostate development (Huang et al., 2007). Recently, we identified HOXB13 in a T7 phage display screen intended to identify proteins that interact with AR (Norris et al., 2009). In this study, we show that HOXB13 is a multifaceted regulator of AR biology, functioning to activate or repress transcription of distinct AR target genes. The complex action of HOXB13 on AR transcriptional activity relates to its ability to both positively and negatively regulate the interaction of AR with chromatin. Importantly, we find that HOXB13 is a critical regulator of the cellular response to androgens.
RESULTS
Isolation of HOXB13 and Characterization of its Interaction with AR
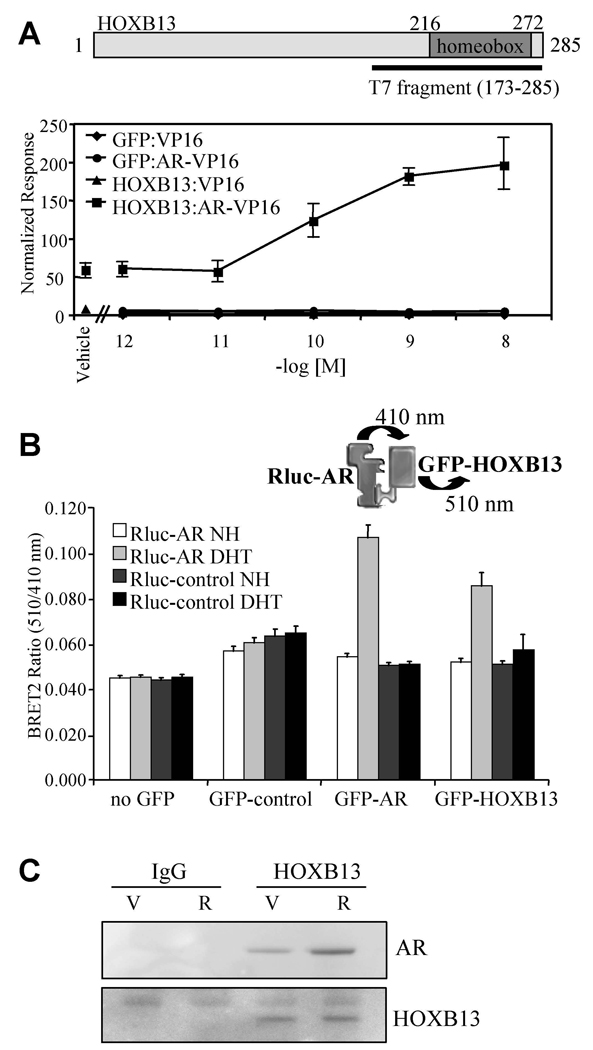
T7 phage display was used to screen cDNA expression libraries from multiple androgen-regulated tissues for proteins that interact with AR (Norris et al., 2009). One of the proteins identified from a human prostate library was HOXB13, a previously characterized AR interacting protein (Jung et al., 2004). A schematic diagram of HOXB13 along with the AR interacting fragment (aa 173–285) isolated from the screen is shown in Figure 1A. The AR interacting domain identified includes the homeobox, a highly conserved DNA binding domain found in the homeodomain gene family of sequence-specific transcription factors. The ability of full-length HOXB13 to interact with AR in cells was first assessed using a mammalian two-hybrid assay (Figure 1A). HOXB13 was found to interact with AR in an R1881 (synthetic AR agonist)-stimulated manner although a significant interaction was also observed with the apo receptor (compare HOXB13:VP16:vehicle with HOXB13:AR-VP16:vehicle).
Figure 1. HOXB13 Interacts with AR in Cells.
(A) Mammalian two-hybrid assay. HepG2 cells were transfected with AR-VP16 or VP16 alone, pCMV-β-gal, 5XGalLUC3 (reporter), and either pM-HOXB13 or pM-GFP (Gal4 DNA binding domain fusion protein constructs). Cells were induced for 48 h with R1881 (as indicated) and assayed for luciferase and β-galactosidase activity. Data is presented as normalized response which was obtained by normalizing luciferase activity with β-galactosidase activity. Error presented as +/− SD of triplicate points.
(B) BRET assay. 293T cells were transfected with pRlucC1-AR (luciferase-AR fusion) or pRluc-C1 (luciferase only) together with either no GFP, empty GFP control, GFP-AR or GFP-HOXB13. GFP-AR was used as a positive control in the dimerization assay. Cells were harvested for analysis following 1 h treatment with DHT (100nM) or vehicle control as indicated (NH; vehicle control, DHT; 5α-dihydrotestosterone). Renilla luciferase acts upon the substrate DeepBlue C to emit light at 410 nm which excites the GFP in close proximity to emit light at 510 nm. The interaction between the luciferase and GFP fusion proteins was measured and expressed as BRET ratio (510nm/410nm). Data shown is the average of 6 independent experiments +/− SEM.
(C) Coimmunoprecipitation. LNCaP cells (100mm dishes) were induced with ligand as indicated (V; vehicle, R; 10nM R1881) for 2 h. Immunoprecipitations were performed using either IgG or anti-HOXB13 antibody. The resultant immunocomplexes were resolved using SDS-PAGE and western blot was performed using either anti-AR or anti-HOXB13 antibodies.
The interaction between AR and HOXB13 was also evaluated using bioluminescence resonance energy transfer (BRET) assay. Since no localization signals are incorporated into the donor/acceptor fusion proteins (Renilla Luciferase and GFP), this assay allows the interaction between AR and HOXB13 to be evaluated in their native compartments. Consistent with the mammalian two-hybrid assay results, HOXB13 and AR were found to interact in a ligand-stimulated manner using the BRET assay (Figure 1B). For both control and comparative purposes, the ability of AR to homodimerize was also evaluated using this assay format. Finally, using a coimmunoprecipitation assay, we showed that HOXB13 and AR could form endogenous complexes (Figure 1C). Cumulatively, these results provide compelling evidence that HOXB13 and AR interact directly with each other in living cells.
HOXB13 Regulates AR Action on Endogenous Target Genes
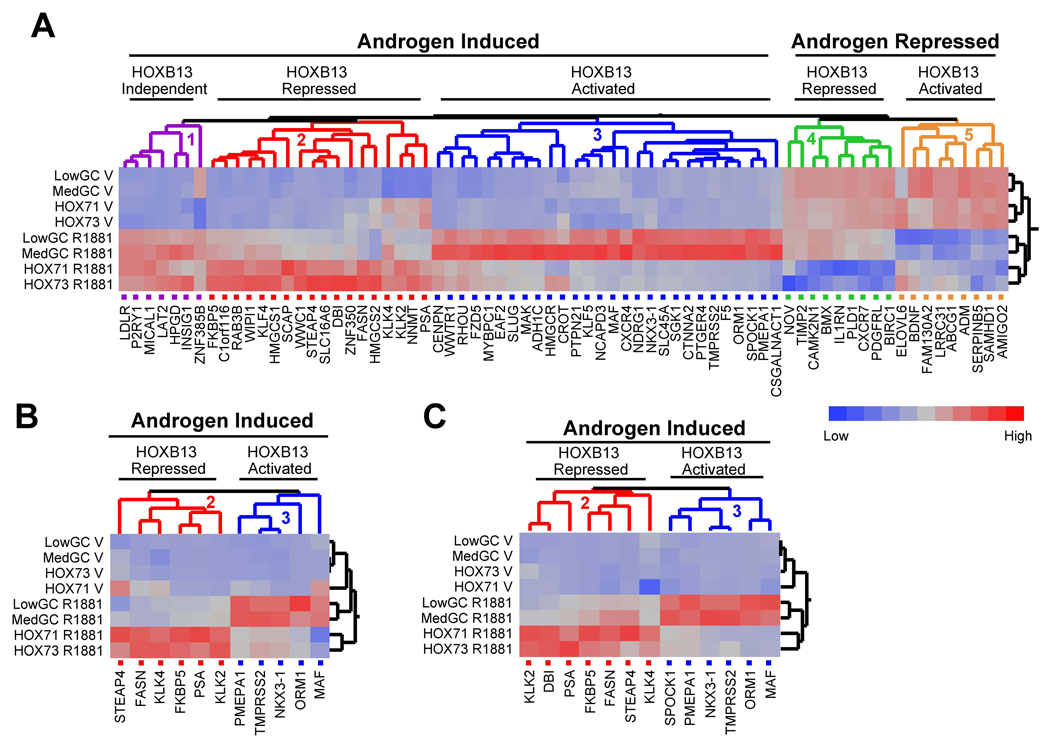
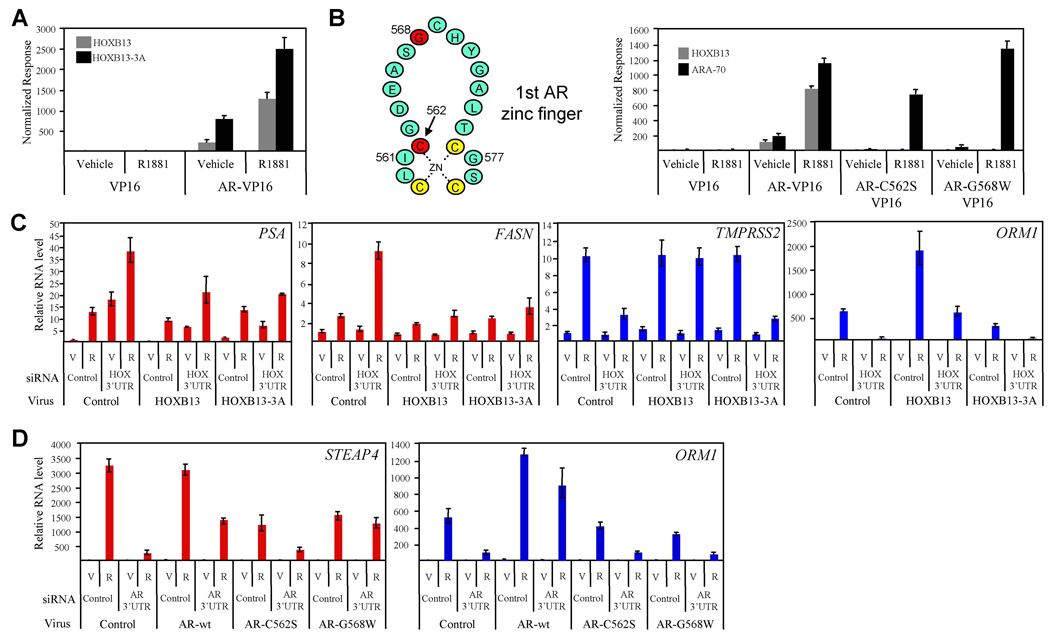
Previously, it has been shown that overexpression of HOXB13 inhibits the transcriptional activity of AR in reporter gene-based assays (Jung et al., 2004). We also found that HOXB13 was a repressor of AR activity when assayed using transiently transfected reporter genes (Figure S1 available with this manuscript online). However, when RNA interference was utilized to specifically suppress endogenous HOXB13 protein levels, a more complex picture emerged. Namely, HOXB13 was found to function as both a positive and a negative regulator of AR target gene transcription. From a microarray analysis performed in both LNCaP and LAPC4 cells (not shown), we selected 71 androgen-regulated genes (53 androgen-activated and 18 androgen-repressed) to generate an AR gene signature (primer sequences for real-time PCR provided in Table S1). The genes included in the signature were either (a) previously identified, well-characterized androgen regulated targets or (b) genes we determined to be induced/repressed by greater than two-fold in androgen treated cells. No other pre-selection criteria were utilized. Using an siRNA approach, we determined that HOXB13 knockdown had a dramatic effect on the expression of the androgen-responsive gene signature in LNCaP cells resulting in at least five distinct phenotypic clusters (Figure 2A). Two sequence-specific siRNAs for both control (LowGC, MedGC) and HOXB13 (HOX71, HOX73) were utilized to minimize the confounding influence of off-target effects on the interpretation of the results. In addition, to confirm the AR-dependence of the genes used for this analysis, we transfected cells with two independent siRNAs targeting AR (AR18, AR19). The detailed results of this study are presented in Table S2. In order to ensure that the siRNAs resulted in the expected reduction in corresponding protein levels, western blot analysis was performed (Figure S2). Cluster one genes (purple) are induced by androgens but are not regulated by HOXB13 to a significant degree. Cluster two genes (red), however, demonstrate a significant increase in androgen-responsiveness upon HOXB13 knockdown suggesting that HOXB13 is functioning as a suppressor of this gene class. Contained within this cluster is a sub-node consisting of four genes, including PSA (KLK3) and other kallikreins (KLK2, KLK4), that demonstrated an increase in both basal and androgen-stimulated transcription with HOXB13 knockdown. Notably, siRNA-mediated knockdown of AR confirmed that the increased basal expression of PSA was entirely receptor-dependent, a result that is in agreement with the repressor function of HOXB13 on this class of genes (Figure S3). Other genes in cluster two that were demonstrated to be negatively regulated by HOXB13 in a robust manner included fatty acid synthase (FASN), FKBP5, and STEAP4. Strikingly, cluster three genes (blue) demonstrated a phenotype opposite to cluster two genes in that they were found to be dependent on HOXB13 for androgen-responsiveness. Thus, HOXB13 acts as a transcriptional coactivator for genes found in cluster three including the well-characterized AR targets TMPRSS2, NKX3.1 and PMEPA1 (TMEPAI).
Figure 2. HOXB13 is a Key Regulator of Endogenous AR Target Gene Transcription.
(A) Dendogram showing expression profiles of endogenous AR target genes (AR gene signature) upon HOXB13 knockdown. Gene expression profiles of 53 genes induced and 13 genes repressed by R1881 treatment (100nM) were generated using LNCaP cells transfected with siRNAs for HOXB13 (HOX71, HOX73) and control (LowGC, MedGC). Following 24 h hormone treatment, RNA was harvested for cDNA production and real-time PCR analysis. The profiles were analyzed with the Ward hierarchical clustering algorithm using standardized data. V; vehicle.
(B) AR gene signature in LAPC4 cells. Experiment was performed same as in (A).
(C) AR gene signature in VCAP cells. Experiment was performed same as in (A).
Interestingly, when this analysis was extended to genes that were repressed by androgens, we found the same disparate regulation. Cluster four genes (green) demonstrated enhanced androgen-mediated repression when HOXB13 protein levels were suppressed (HOXB13 inhibits androgen-mediated repression) while cluster five genes (orange) demonstrated HOXB13-dependent repression (HOXB13 facilitates androgen-mediated repression). This complex regulation of the androgen signaling axis was not limited to LNCaP cells as we observed the same activity when an analogous study was performed in two additional prostate cancer cell lines, LAPC4 (Figure 2B) and VCAP (Figure 2C). Furthermore, the regulation of AR transcriptional activity by HOXB13 was observed at both high and low hormone levels (Figure S4). Cumulatively, these results demonstrate that HOXB13 is a bifunctional regulator of AR transcriptional activity, demonstrating the hallmarks of both an activator and a repressor.
HOXB13 Regulates the Interaction of AR and its Associated Coregulators with Chromatin
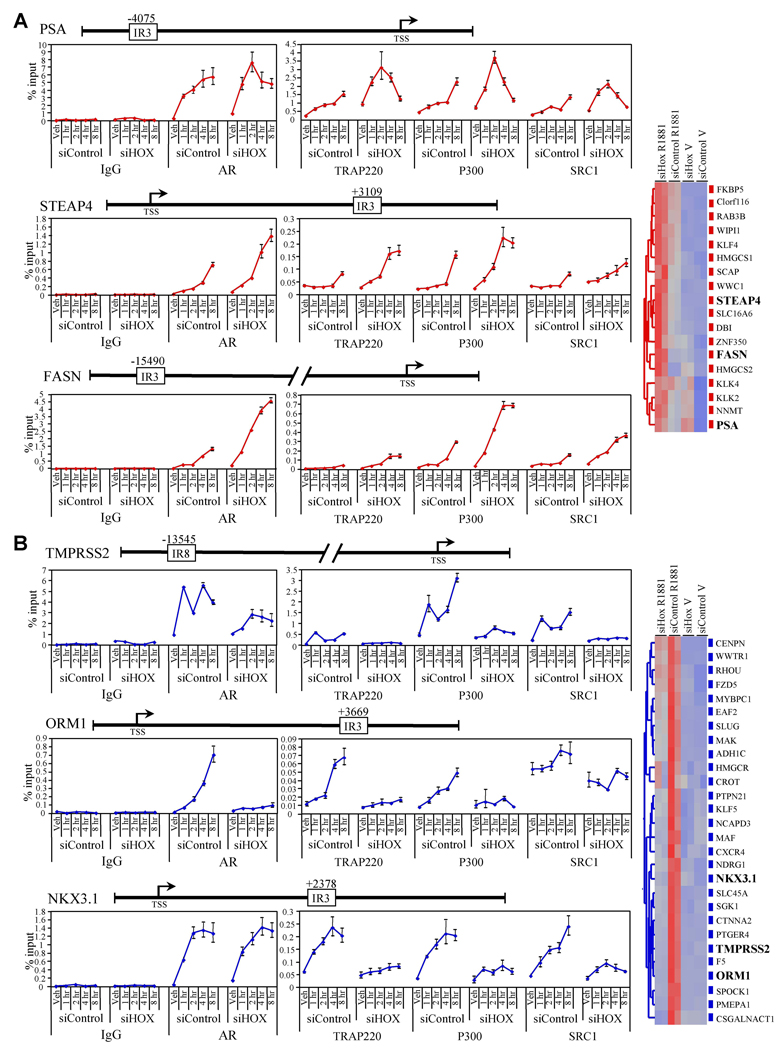
Given the robust differential regulation of endogenous AR target gene transcription by HOXB13, we explored its role in regulating the interaction of AR with chromatin at distinct target gene promoters. Using a combination of bioinformatics (search for inverted repeat 3 (IR3; AGAACAnnnTGTTCT) or other described androgen response elements (Wang et al., 2007)) and chromatin immunoprecipitation (ChIP) scanning, we mapped the specific AR interacting regions within the enhancers of several gene promoters on which HOXB13 functioned as a repressor (Figure 3A, cluster 2 genes) or an activator (Figure 3B, cluster 3 genes). The location of the AR-interacting sequences identified, along with those previously reported in the literature (PSA (distal enhancer), TMPRSS2) (Cleutjens et al., 1997; Wang et al., 2007), are indicated in the gene illustrations relative to the transcriptional start site as annotated by the USCS genome browser (http://genome.ucsc.edu). Interestingly, the AR-binding regions for STEAP4, ORM1, and NKX3.1 were found in intron 1, 3’ regulatory, and 3’UTR respectively. The AR binding region for FASN was found to reside in the 5’ regulatory region, a location similar to those previously identified for PSA and TMPRSS2.
Figure 3. HOXB13 Regulates the DNA Binding Activity of AR.
(A) Time course ChIP assay for cluster 2 genes. LNCaP cells (150mm dishes) were transfected with either siControl (LowGC) or siHOXB13 (siHOX71) for 48 h prior to the addition of R1881 (100nM). Following hormone addition (1,2,4, and 8 h), cells were fixed with formaldehyde and sheared chromatin was immunoprecipitated with the indicated antibodies (IgG, AR, TRAP220, P300, SRC1). Real-time PCR was used to assess the amount of DNA immunoprecipitated in response to hormone treatment, presented as a percentage of input chromatin. The position of the AR binding sites (IR3 or IR8) identified by bioinformatic analysis in relation to the transcription start site (TSS) is provided in the gene structure illustrations. Error presented as +/− SD of triplicate points. Veh; vehicle.
(B) Time course ChIP assay for cluster 3 genes. Experiments were performed same as in (A). Data for (A) and (B) are colored according to gene cluster as presented in Figure 2.
In a time course ChIP analysis, it was demonstrated that HOXB13 knockdown increased the R1881-mediated recruitment of AR and several of its attendant coregulatory proteins (TRAP220, P300, SRC1) to the PSA (distal), STEAP4, and FASN enhancers (Figure 3A). The increase in coregulatory protein recruitment was not due to a change in coregulatory protein expression levels (Figure S5). HOXB13 RNA interference also had a significant effect on basal AR and coregulator (particularly TRAP220) recruitment (increase of approximately 5 fold) to the PSA enhancer, mimicking the effect of HOXB13 knockdown on PSA gene expression (Figure 2 and S3). Interestingly, HOXB13 was also found to regulate the recruitment of AR and AR-coregulators to these genes in a temporal manner. This was particularly evident for the STEAP4 and FASN enhancers where reduced levels of HOXB13 decreased the lag time between the addition of hormone and the appearance of AR and AR coregulators at the promoter. In addition to the distal enhancer (located approximately −4200 from the transcription start site), the PSA gene has also been shown to contain a second proximal enhancer (located from −170 to −400) (Cleutjens et al., 1997). Similar to what was observed for the distal enhancer (Figure 3A), we demonstrated that HOXB13 knockdown led to a modest increase in R1881-mediated recruitment of AR to the proximal enhancer (Figure S6). These data are consistent with the target gene transcription studies presented in Figure 2 and provide confirmation of the functionality of the AR binding regions that we identified in these genes.
To assess the role of HOXB13 in the regulation of cluster three genes, we performed a time course ChIP analysis as above (Figure 3B). Contrary to the results found for cluster two genes, knockdown of HOXB13 led to a striking reduction of both AR and AR-coregulator recruitment to the TMPRSS2 and ORM1 enhancers in an R1881-dependent manner. Interestingly, a different phenotype emerged when this analysis was performed for the NKX3.1 gene. Reduction in HOXB13 protein levels had a minimal effect on AR recruitment to the NKX3.1 enhancer but had a significant effect on AR-coregulator recruitment. Thus, for cluster three genes, HOXB13 appears to act as a licensing factor not only for AR and AR-coregulator recruitment to chromatin (TMPRSS2, ORM1) but also as a licensing factor for coregulator recruitment to AR itself (NKX3.1). It should also be noted that although HOXB13 knockdown affects AR recruitment to the TMPRSS2 enhancer, it has a more pronounced effect on coregulator recruitment. As with cluster two genes, the HOXB13 dependent regulation of AR and AR-coregulator recruitment to cluster three gene enhancers is consistent with the transcriptional studies presented in Figure 2.
HOXB13 Regulates AR Target Gene Enhancer activity in Both a Positive and Negative Manner
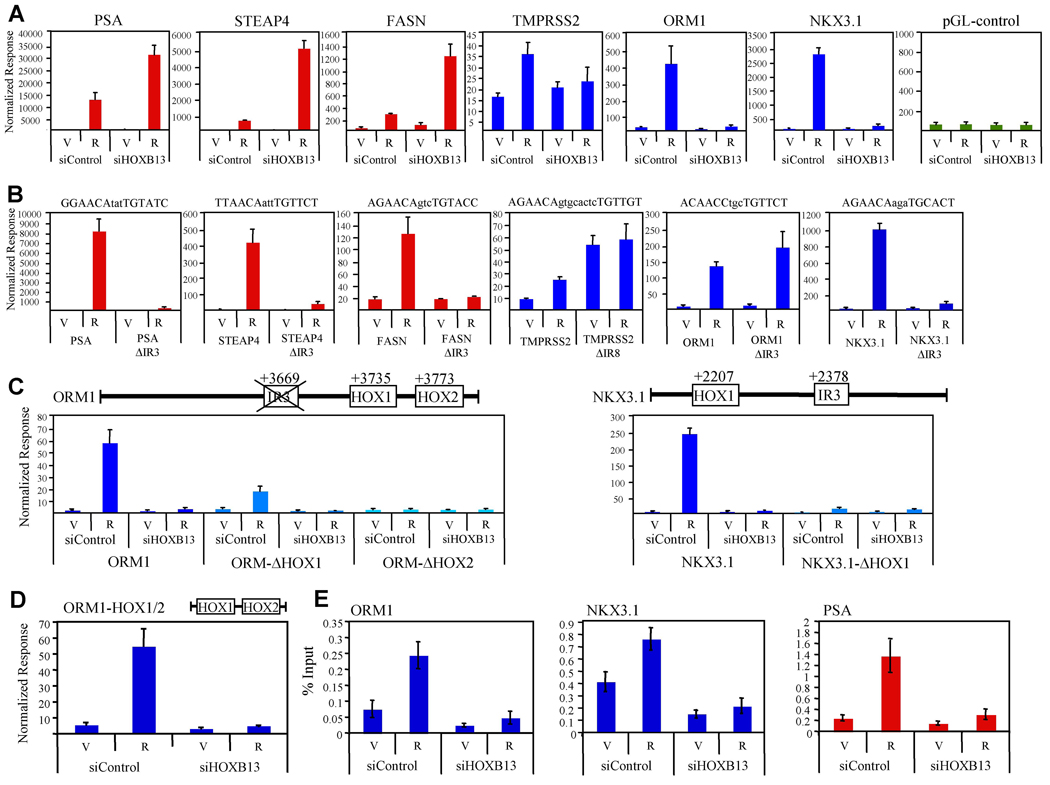
To define the mechanism(s) by which HOXB13 regulates AR target gene transcription, each of the six HOXB13-regulated gene enhancer regions (300–800 bp) described above (PSA, FASN, STEAP4, TMPRSS2, ORM1, NKX3.1) were cloned into a reporter gene containing a minimal promoter. The androgen-dependent regulation of these reporter genes was then assessed in both the presence and absence of a siRNA targeting HOXB13 (Figure 4A). When HOXB13 proteins levels are reduced in LNCaP cells, androgen-dependent regulation of cluster 2 gene enhancers (PSA, FASN, STEAP4) is strongly enhanced. An analogous study using cluster 3 gene enhancers (TMPRSS2, ORM1, NKX3.1) demonstrated a profound inhibition of R1881-mediated transcription upon loss of HOXB13. These results are strikingly consistent with those observed for the corresponding endogenous genes (Figure 2), suggesting that the enhancer regions identified are at least partially responsible for both the AR- and HOXB13-dependent regulation of these genes. Furthermore, deletion of the in silco-defined AR binding sites (IR3 or IR8) within these target gene reporter constructs confirms that all the sites identified, except for the one found in ORM1, are functional (Figure 4B). Mutation of other potential AR binding sequences within the ORM1 enhancer did not result in the identification of a functional ARE (androgen response element) suggesting that AR does not utilize a canonical ARE for its regulation of this enhancer (not shown). Finally, we utilized an siRNA targeting AR to confirm the AR dependence of these reporter genes (Figure S7).
Figure 4. Regulation of AR Target Gene Enhancers by HOXB13.
(A) HOXB13 is both a coactivator and corepressor. LNCaP cells were transfected with indicated siRNA for 24 h. Cells were then transfected with pCMV-β-Gal and indicated reporter gene (PSA, FASN, STEAP4, TMPRSS2, ORM1, NKX3.1) for 24 h prior to the addition of R1881 (100nM). Empty reporter vector (pGL-control) is included as control. Following 24 h hormone addition, cell lysates were assayed for luciferase and β-galactosidase activity and data was normalized as in Figure 1A. Data is colored according to gene cluster as presented in Figure 2 (red; cluster 2, blue; cluster 3). Error presented as +/− SD of triplicate points. V; vehicle, R; R1881.
(B) Functional characterization of the AR binding sites identified within the promoters of androgen-responsive genes. Indicated above each panel is the potential AR binding sequence that was deleted to create the ΔIR3 or ΔIR8 mutant for each reporter gene. LNCaP cells were then transfected with the wildtype reporter gene or its corresponding mutant for 24 h before hormone addition (100nM R1881). Following 24 h incubation, cell lysates were assayed for luciferease and β-galacatosidase activity as detailed in (A).
(C) Characterization of HOX DNA binding sites in androgen responsive genes. Indicated above each panel is a gene diagram showing the relative position of potential HOX DNA binding sites in both the ORM1 and NKX3.1 gene enhancers. LNCaP cells were transfected with the indicated reporter gene and assayed as in (A).
(D) LNCaP cells were transfected as in (A) with an ORM1 reporter gene containing only the HOX1 and HOX2 binding sites (ORM1-HOX1/2). Error presented as +/− SD of triplicate points.
(E) HOXB13 is recruited to chromatin. LNCaP cells (150mm dishes) were transfected with either siControl (LowGC) or siHOXB13 (HOX71) for 48 h prior to the addition of R1881 (10nM). Following hormone addition (4 h) cells were fixed with formaldehyde and sheared chromatin was immunoprecipitated with IgG control or anti-HOXB13 antibody (H80, Santa Cruz Biotechnologies). Real-time PCR was used to assess the amount of DNA immunoprecipitated in response to hormone treatment, presented as a percentage of input chromatin. Error presented as +/− SD of triplicate points.
Given that the androgen-responsiveness of cluster 3 genes was found to be HOXB13-dependent, we reasoned that their corresponding enhancers might contain DNA binding sites for HOXB13. To explore this possibility, we used deletion analysis to systematically mutate potential HOX binding sites within the ORM1 and NKX3.1 enhancers (Figure 4C). The results of these studies reveal that within that ORM1 enhancer, there are two potential HOX binding sequences (TTTAC; ORM-HOX1 and ATAAA; ORM-HOX2) that are critical for androgen responsiveness. A similar analysis of the NKX3.1 promoter revealed that the potential HOX binding sequence GTAAA (NKX3.1-HOX1) is required for full AR activity.
The unexpected absence of a canonical ARE within the ORM1 enhancer, combined with the finding that HOXB13 and AR can interact directly with one another, prompted us to explore the possibility that AR was recruited to the ORM1 enhancer by its direct association with HOXB13. To address this question, we utilized a minimal ORM1 enhancer fragment containing only the HOX1 and HOX2 binding sites (ORM1-HOX1/2) to show that AR can stimulate transcription from this reporter gene in the absence of any ARE (Figure 4D).
Next, we utilized a ChIP assay to investigate whether endogenous HOXB13 can interact with the potential HOXB13 binding sites identified by the deletion analysis above. As shown in Figure 4E, we found that HOXB13 is recruited to both the ORM1 and NKX3.1 (cluster 3 genes) enhancers in an androgen-enhanced manner. Furthermore, these studies revealed significant ligand-independent interaction of HOXB13 with the ORM1 and NKX3.1 enhancers (compare siControl:vehicle with siHOXB13:vehicle). In addition to the cluster 3 gene enhancers, we demonstrated also that HOXB13 interacted with the PSA (cluster 2 gene) distal enhancer (Figure 4D). Cumulatively, these results show that HOXB13 is recruited to both cluster 2 and cluster 3 gene enhancers in an androgen-stimulated manner. Next, we focused on defining the roles of the DNA binding domains of both HOXB13 and AR in mediating the androgen-responsiveness of cluster two and cluster three genes.
Role of HOXB13 and AR DNA Binding Domains in Androgen-regulated Target Gene Transcription
To define the role of the HOXB13 homeobox in modulating AR transcriptional activity, we first tested the ability of a HOXB13 DNA binding mutant (HOXB13WFQ>AAA , HOXB13-3A) (Suzuki et al., 2003) to interact with AR in a mammalian two-hybrid assay. Interestingly, point mutations in the homeobox known to prevent HOXB13 binding to DNA did not affect its interaction with AR (Figure 5A). In contrast, a comparable study using DBD mutants of AR (AR-C562S, AR-G568W) demonstrated the importance of the AR-DBD in mediating the interaction between AR and HOXB13 (Figure 5B). Furthermore, in order to confirm that the AR mutations do not effect global receptor conformation, we demonstrated that the previously characterized AR coregulator, ARA70 (Yeh and Chang, 1996), interacted with both wild-type AR and the AR DBD mutants.
Figure 5. HOXB13 DNA Binding is Required for Coactivator but not Corepressor Function.
(A) HOXB13 DNA binding mutant interacts with AR. A Mammalian two-hybrid assay was performed as in Figure 1A using wildtype HOXB13 (pM-HOXB13) or a DNA binding mutant of HOXB13 (pM-HOXB13-3A). Error presented as +/− SD of triplicate points.
(B) AR DNA binding domain mutants do not interact with HOXB13. Schematic representation of the first zinc finger in the AR DNA binding domain is shown with mutant residues in red. Mammalian two-hybrid assay was performed using wildtype HOXB13 (pM-HOXB13) and wildtype AR (AR-VP16) or mutant AR (AR-C562S-VP16, AR-G568W-VP16).
(C) HOXB13 complementation assay. Retrovirus was used to generate stable LNCaP cell lines expressing HOXB13, HOXB13-3A, or control (XIN). Cells were transfected with siRNA targeting the 3’UTR of endogenous HOXB13 mRNA (targets endogenous HOXB13 and not viral-expressed HOXB13) or control siRNA. Following 48 h incubation, cells were treated with vehicle (V) or R1881 (R, 100nM) for 24 h. Real-time PCR was used to assess androgen-responsiveness of indicated AR target genes. Data are colored according to gene cluster as presented in Figure 2. Error presented as +/− SD of triplicate points.
(D) AR complementation assay. LNCaP cells were transfected with control siRNA or siRNA targeting the 3’UTR of AR for 24 h. Cells were then transiently infected with retroviruses expressing wt-AR, AR-C562S, AR-G568W, or control (pMIG). Following 48 h incubation, cells were treated with vehicle (V) or R1881 (R, 100 nM) for 24 h. Real-time PCR analysis was performed as in (C).
Using retroviruses expressing either wild-type (wt)-HOXB13 or HOXB13-3A, we demonstrated that the repressor function of HOXB13, as manifest on the androgen-responsiveness of cluster 2 genes (PSA, FASN), does not require its previously defined DNA binding activity (Figure 5C). Specifically, we utilized an siRNA that targets the 3’UTR of the endogenous HOXB13 mRNA (siHOX-3’UTR) but not the virally expressed HOXB13 mRNA to perform complementation studies with either virally expressed wt-HOXB13 or HOXB13-3A. Western immunoblot was used to demonstrate the specificity of the HOXB13 3’UTR in targeting only endogenous HOXB13 (Figure S8). These studies demonstrated that wt-HOXB13 and HOXB13-3A both have repressor activity on the PSA and FASN genes (compare XIN: HOX 3’UTR: R1881 with HOXB13: HOX 3’UTR: R1881 or HOXB13-3A: HOX 3’UTR: R1881). Interestingly, these results combined with the finding that HOXB13 is recruited to the PSA distal enhancer in an androgen-stimulated manner (Figure 4E) suggests that HOXB13 is recruited to and represses cluster 2 genes through its direct association with AR and not through its own DNA binding activity.
We next performed a similar study evaluating the role of the homeobox (DNA-binding domain) in mediating the activation function of HOXB13 on the androgen-responsiveness of cluster 3 genes (TMPRSS2, ORM1). In this study, we found that HOXB13 DNA binding was absolutely required for the androgen-regulated expression of both the TMPRSS2 and ORM1 genes (compare XIN: HOX 3’UTR: R1881 with HOXB13: HOX 3’UTR: R1881 or HOXB13-3A: HOX 3’UTR: R1881) (Figure 5C). Cumulatively, these data demonstrate that the activator and repressor functions of HOXB13, with respect to androgen regulated genes, can be separated based on the DNA binding activity of HOXB13 and furthermore provides additional evidence that the HOX binding sites identified within the ORM1 enhancer interact directly with HOXB13.
The ability of HOXB13 to interact with the AR DBD coupled with the results from the ChIP analysis (Figure 3A) suggested that HOXB13 functions as a repressor by interfering with the ability of AR to interact with its cognate DNA response elements. We infer also from these results that HOXB13 can function as a “coregulator” by recruiting AR to target gene promoters and conferring upon them the ability to be regulated by androgens. To test this alternate mode of regulation, we utilized an siRNA that specifically targets the 3’UTR of the endogenous AR mRNA (siAR-3’UTR) but not the virally expressed AR mRNA to perform complementation studies with either virally expressed wt-AR, AR-C562S, or AR-G568W (Figure 5D). Although both AR mutations prohibit the binding of HOXB13 (Figure 5B), only the AR-C562S mutation has been shown to prevent AR from activating reporter genes containing a bona fide ARE (Ikonen et al., 1994; Lobaccaro et al., 1999). This demonstrates that the AR-G568W mutation is still functional with respect to DNA binding activity. Western immunoblot analysis was used to demonstrate the specificity of the siAR 3’UTR in targeting only endogenous AR and also provided evidence that the AR protein levels in the complementation study are comparable to endogenous levels (Figure S9). The results of this analysis demonstrated that as expected the ARE-containing STEAP4 gene can be complimented by both wt-AR and AR-G568W but not by AR-C562S (compare AR-wt: AR 3’UTR: R1881 with AR-C562S: AR 3’UTR: R1881 and AR-G568W: AR 3’UTR: R1881) (Figure 5D). In contrast, the ORM1 gene can only be complimented by wt-AR and not by AR-C562S or AR-G568W. A model describing the interaction between the AR mutants, DNA, and HOXB13 is shown in Figure S10. These results provide strong evidence that there are no canonical AR binding sites within the ORM1 gene and that AR most likely regulates this gene through its interaction with HOXB13.
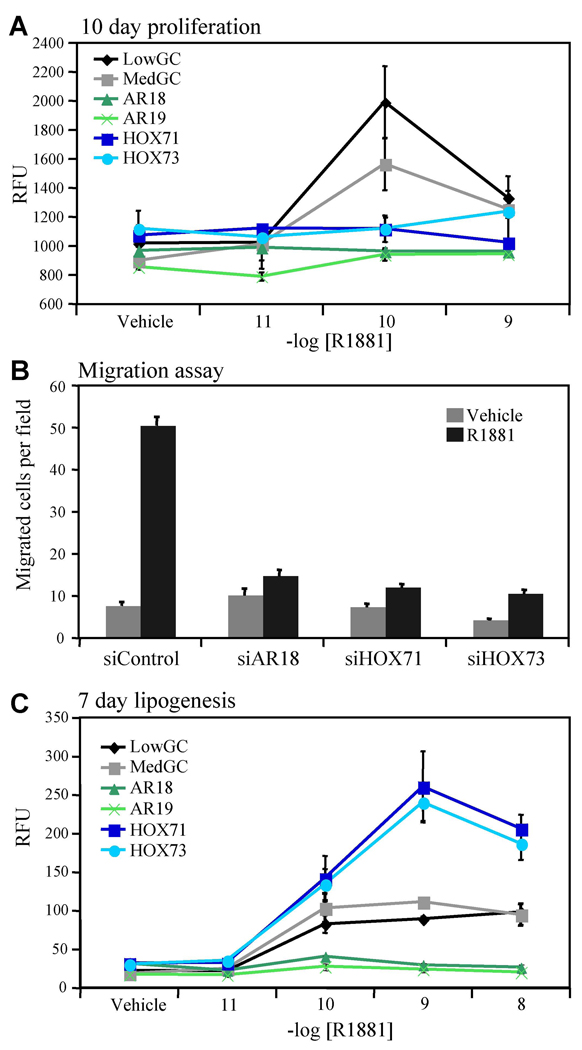
HOXB13 Regulates Androgen-dependent Proliferation, Migration, and Lipogenesis
With the goal of gaining a better understanding of the impact of HOXB13 on physiologically/pathologically relevant AR-regulated processes, we examined its role in androgen-mediated proliferation, migration, and lipogenesis. For the proliferation studies, LNCaP cells were transfected with two sequence-specific siRNAs targeting HOXB13, two siRNAs targeting AR and two control siRNAs. Surprisingly, reduction of HOXB13 protein levels resulted in a near complete loss of cell growth in response to androgen treatment suggesting that HOXB13 is required for AR-stimulated proliferation of LNCaP cells (Figure 6A). Similar to what was observed in the proliferation study, HOXB13 was also found to be critical for androgen-stimulated cell migration (Figure 6B). In addition to stimulating cell growth, androgens also promote de novo synthesis of intracellular lipids (Swinnen et al., 1996). Notably, HOXB13 suppression increased androgen-stimulated total lipid accumulation when compared to control cells whereas knockdown of AR resulted in a complete loss of lipid accumulation, confirming the central role of the receptor in this androgen-regulated process (Figure 6C). Taken together, these results demonstrate that the multifaceted regulation of AR target gene transcription by HOXB13 (activator and repressor) is also manifest in its complex regulation of AR biology.
Figure 6. HOXB13 Determines the Cellular Response to Androgens.
(A) Proliferation assay. LNCaP cells were transfected with indicated siRNA and seeded for 3 days in medium containing charcoal stripped serum. On days 3, 6, and 9 the cells were treated with indicated concentration of R1881. Proliferation was determined by measuring total cellular DNA content on day 10. Data is presented as +/− SD of triplicate wells.
(B) Migration assay. LNCaP cells were transfected with indicated siRNA. Two days after transfection, cells were treated with R1881 (10nM) or vehicle in serum-free media. 24 h later, cells were plated in triplicate in the top of a transwell and allowed to migrate across a fibronectin-coated membrane toward a serum gradient. After 18 h, migrated cells were stained and three high-powered fields were counted per transwell. Data is presented as the average number of cells migrated +/− SD for three fields of a representative experiment.
(C) Lipogenesis assay. LNCaP cells were transfected with indicated siRNA and seeded for 3 days in medium containing charcoal stripped serum. On days 3 and 6 the cells were induced with the indicated concentration of R1881. Lipid accumulation was measured using AdipoRed. Data is presented as +/− SD of triplicate wells.
DISCUSSION
We identified HOXB13 using a screen designed to identify proteins that interact directly with and modulate AR function in cells (Norris et al., 2009). Characterization of this homeodomain protein revealed that it plays a central role in AR signaling, acting as a repressor on some gene promoters and an obligate coactivator on others. Unlike a number of AR coregulators, HOXB13 lacks enzymatic activity and appears to impact transcription by positively or negatively regulating (a) the recruitment of AR to target genes and (b) the recruitment of coregulators to DNA-bound AR. Thus, HOXB13 functions as a key licensing factor for both AR and AR-coregulator recruitment to chromatin.
Early studies of NR coregulators led to their classification as either coactivators or corepressors. Recently, our understanding of coregulator function has evolved and it is now apparent that their activity is context dependent (Li et al., 2002; O'Malley, 2007). For example, NCoR, a well-characterized NR co-repressor, has also been shown to have coactivator activity (Peterson et al., 2007). In this regard, the majority of studies ascribing a regulatory role to NR-interacting proteins rely on either transfected reporter genes or on the analysis of a few representative target genes. The data presented here demonstrates that the AR coregulator HOXB13 can function to efficiently repress or activate a significant number of endogenous AR target genes. Importantly, the differential effects of HOXB13 on target gene transcription are also manifest in complex AR biology.
Previous studies using overexpression strategies showed that HOXB13 was a repressor of prostate cancer cell proliferation (Jung et al., 2004). By contrast, in this study, we showed that prostate cancer cell growth was HOXB13-dependent. These discrepancies may be due to the different experimental strategies (overexpression vs. siRNA) that were used to assess the role of HOXB13 in androgen-mediated proliferation. Interestingly, a recent study demonstrated that HOXB13 was a pro-proliferative and prosurvival factor in ER positive ovarian cancer cells (Miao et al., 2007) supporting our finding that HOXB13 is a pro-proliferative factor for prostate cancer cells in response to androgens. Furthermore, we recently reported that serum- and glucocorticoid-regulated kinase 1 (SGK1) was an androgen-regulated gene in LNCaP cells and that it was required for androgen-responsive cell growth (Sherk et al., 2008). Consistent with this observation, we found that SGK1 was entirely dependent on HOXB13 for androgen-stimulated expression (Figure 2). In normal, differentiated, prostatic epithelial cells androgens regulate the de novo synthesis of neutral lipids. One of the critical enzymes in this pathway is FASN, a polypeptide that catalyzes the synthesis of fatty acids from acetyl-CoA and malonyl-CoA (Swinnen et al., 1997). Our results showing that the FASN gene is significantly repressed by HOXB13 is consistent with the observation that HOXB13 suppresses fatty acid accumulation. Cumulatively, these data support a model whereby HOXB13 is a key regulator of the balance between cell growth (proliferation) and a more differentiated state (lipogenesis). Furthermore, it demonstrates that HOXB13 regulates AR target gene transcription in a highly coordinated manner to mediate the biological actions of androgens.
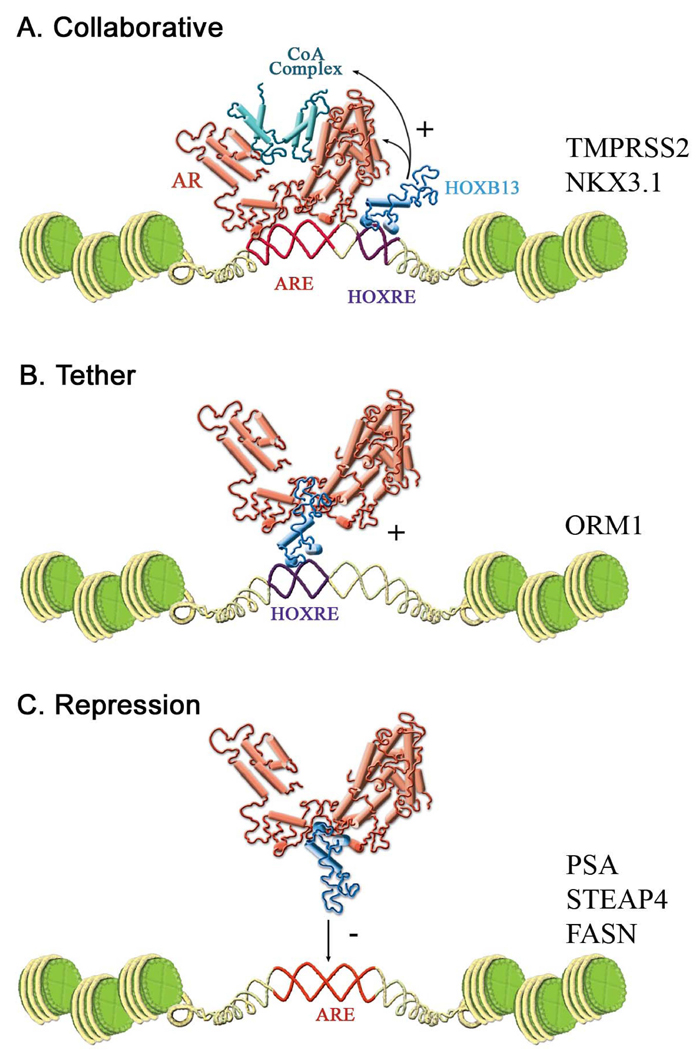
The precise mechanism by which HOXB13 regulates the loading of AR and AR-coregulatory complexes onto chromatin awaits further study. Here, we propose the following models to explain the multifaceted regulation of the AR signaling axis by HOXB13 (Figure 7). Androgen-regulated genes that are HOXB13-dependent (cluster 3 genes) can generally be subdivided into two distinct classes. In the “collaborative model” of gene activation, represented by the TMPRSS2 and NKX3.1 genes, DNA binding sites for both AR and HOXB13 are present (Figure 7A). These genes each have validated AREs within their respective enhancers (Figure 4B). Additionally, the recruitment of HOXB13 to the NKX3.1 gene enhancer (Figure 4E) combined with the results from the deletion analysis of the NKX3.1 gene enhancer (Figure 4C) indicates that it also contains a functional HOXB13 binding site(s). We did not attempt to map the HOXB13 binding site/s within the TMPRSS2 enhancer given that it was not very active in transient transfection assays; however, the dependence of androgen-stimulated TMPRSS2 gene transcription on the DNA binding activity of HOXB13 (Figure 5C) makes it likely that this gene also contains functional HOXB13 binding site(s). One notable difference between the TMPRSS2 and NKX3.1 genes is that on the latter HOXB13 does not appear to affect AR recruitment (Figure 3B).
Figure 7. Models of AR Target Gene Regulation by HOXB13.
(A) HOXB13 is a licensing factor for AR and AR coregulators (collaborative model). HOXB13 is required for robust recruitment of AR coregulators (TMPRSS2, NKX3.1) and in some instances AR (TMPRSS2) to AREs. The DNA binding function of HOXB13 is required for this activity.
(B) Activation function of HOXB13 (tether model). AR interacts directly with HOXB13 to activate target genes containing HOXB13 binding sites (ORM1). The DNA binding function of HOXB13 is required for AR activity.
(C) Repressor function of HOXB13. HOXB13 interacts with the AR DNA binding domain weakening the association of AR with chromatin. The DNA binding activity of HOXB13 is not required for repression.
The ORM1 gene represents a second class of androgen-stimulated genes that are HOXB13 dependent. In this “tether model” of gene activation (Figure 7B), AR is recruited to chromatin in the absence of a classical ARE. Multiple lines of evidence support this model. First, no ARE could be identified in the cloned ORM1 enhancer region even though it was induced by androgens. Second, a DNA binding domain mutant of AR (AR-G568W) that retained the ability to activate target genes containing an ARE but lost the ability to interact with HOXB13 does not activate the ORM1 gene (Figure 5D). Finally, the identification of functional HOX binding sites (ORM1-HOX1, ORM1-HOX2) within the ORM1 enhancer and the dependence on these sites for full androgen-responsiveness (Figure 4C) provides compelling evidence that AR activates this promoter through its direct interaction with HOXB13. The ability of AR to regulate transcription in a non-classical manner through its interaction with DNA-bound transcription factors (AP-1, NFk-B) has been reported previously (Aarnisalo et al., 1999; Lobaccaro et al., 1999). However, we believe that the tethering mechanism proposed here to explain how HOX proteins regulate gene transcription has not been contemplated previously.
The “repressor model” of HOXB13 regulation is illustrated by the ARE containing genes PSA, STEAP4, and FASN (Figure 7C). In this study, we have shown that HOXB13 interacts with the AR DBD suggesting that HOXB13 can in some way interfere with the DNA binding activity of AR. Interestingly, HOXB13 did not repress all cluster two genes to the same extent as might be expected if the repressor function of HOXB13 can be attributed solely to its ability to physically preclude DNA-receptor interactions. One explanation that may account for this variability is that AR can interact with different AREs in slightly different orientations allowing access of AR-HOXB13 complexes to some gene enhancers but not others. The affinity of AR for different AREs may also play a role in specifying the repressor activity of HOXB13 through competition for the AR-DBD. Finally, it is likely that the promoter context will have a role in regulating the activity of HOXB13-AR complexes.
In summary, the data presented here highlights a critical role for HOXB13 in regulating AR biology in cancer cells. Future studies will examine the regulation of AR by HOXB13 in both developmental and adult tissues. Interestingly, both HOXB13 and AR utilize their respective DNA binding domains to mediate the interaction with each other. Given the highly conserved nature of both the homeobox and the NR-DBD, it appears likely that these two classes of sequence-specific transcription factors may have far-reaching regulatory impacts on each others activity. Finally, the observation that HOXB13 is upregulated in some cancers (Lopez et al., 2006; Yamashita et al., 2006) coupled with the finding that it is a critical determinant for prostate cancer cell growth suggests that targeting HOXB13 or the HOXB13:AR interface may have therapeutic utility.
EXPERIMENTAL PROCEDURES
Chemicals and Plasmids
R1881 was purchased from PerkinElmer (Waltham, MA). DHT was purchased from Steraloids (Newport, RI). HOXB13 (F9, H80) and GAPDH (0441) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). AR-VP16, 5xGal4Luc3, MMTV-Luc, PSA-Luc, and pCMV-βGal were described previously (Chang et al., 2005). pSG5-AR was a gift from T. Willson (GlaxoSmithKline, Research Triangle Park, NC). pMIG (retrovirus vector) was a gift from J. Rathmell (Duke University, Durham, NC). Additional details for chemicals and plasmids can be found in the supplemental data.
Cell Culture, Transfection Assays, and Coimmunoprecipitation
LNCaP cells were maintained in RPMI supplemented with 10% FCS (Invitrogen). HepG2, VCAP, and 293T cells were maintained in DMEM supplemented with 10% FCS. LAPC4 cells were maintained in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 15% FCS plus 0.1 nM R1881. Additional details for the transfection, proliferation, migration, lipogenesis, and coimmunoprecipitation assays can be found in the supplemental data. siRNA sequcences can be found in Table S1.
Bioluminescence Resonance Energy Transfer (BRET)
293T cells were seeded in 24-well plates overnight and transfect with pRlucC1-AR or pRluc-C1 together with either no GFP, empty GFP control, GFP-AR or GFP-hoxb13. Additional details of the BRET assay can be found in the supplemental data.
RNA Isolation and Real-Time PCR
RNA was isolated using the Aurum™ total RNA isolation kit (Bio-Rad, Hercules, CA). RNA (1 µg) was reverse transcribed using the Bio-Rad iScript cDNA synthesis kit. Real-time PCR was performed using the Applied Biosystems (Foster City, CA) 7300 instrument and iQ SYBR Green Supermix (Bio-Rad). GAPDH expression was used to normalize all real-time data. Sequences for gene-specific primers can be found in Table S1.
Chromatin Immunoprecipitation Assay
Details for the ChIP assay can be found in the Supplemental Data. Real-time primer sequences for the ChIP assay can be found in Table S1.
Data Analysis
For AR gene signature, the data was first normalized to the LowGC vehicle control within each gene. To avoid signal strength bias, the data was then standardized using the following equation:
where X is the normalized signal, μ is the average signal for all conditions within a gene and σ is the standard deviation for all conditions within a gene. The data was then clustered by the Ward hierarchical clustering method using JMP (SAS, Cary, NC). The hierarchical cluster dendrogram was ordered by the first principle component and 5 clusters were selected.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by a grant to D.P.M. from the NIH (CA139818).
Abbreviations
- AR
androgen receptor
- ER
estrogen receptor
- NR
nuclear receptor
- BRET
bioluminescence resonance energy transfer
- ARE
androgen response element
- HOXRE
HOXB13 response element
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aarnisalo P, Santti H, Poukka H, Palvimo JJ, Janne OA. Transcription activating and repressing functions of the androgen receptor are differentially influenced by mutations in the deoxyribonucleic acid-binding domain. Endocrinology. 1999;140:3097–3105. doi: 10.1210/endo.140.7.6792. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol. Cell. Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Abdo J, Hartney T, McDonnell DP. Development of peptide antagonists for the androgen receptor using combinatorial peptide phage display. Mol. Endocrinol. 2005;19:2478–2490. doi: 10.1210/me.2005-0072. [DOI] [PubMed] [Google Scholar]
- Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol. Endocrinol. 1997;11:148–161. doi: 10.1210/mend.11.2.9883. [DOI] [PubMed] [Google Scholar]
- Huang L, Pu Y, Hepps D, Danielpour D, Prins GS. Posterior Hox gene expression and differential androgen regulation in the developing and adult rat prostate lobes. Endocrinology. 2007;148:1235–1245. doi: 10.1210/en.2006-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30:965–979. doi: 10.1002/bies.20823. [DOI] [PubMed] [Google Scholar]
- Ikonen T, Palvimo JJ, Kallio PJ, Reinikainen P, Janne OA. Stimulation of androgen-regulated transactivation by modulators of protein phosphorylation. Endocrinology. 1994;135:1359–1366. doi: 10.1210/endo.135.4.7925097. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J. Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- Jung C, Kim RS, Zhang HJ, Lee SJ, Jeng MH. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004;64:9185–9192. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J. Biol. Chem. 2002;277:26031–26035. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kimbrel EA, Kenan DJ, McDonnell DP. Direct interactions between corepressors and coactivators permit the integration of nuclear receptor-mediated repression and activation. Mol. Endocrinol. 2002;16:1482–1491. doi: 10.1210/mend.16.7.0860. [DOI] [PubMed] [Google Scholar]
- Lobaccaro JM, Poujol N, Terouanne B, Georget V, Fabre S, Lumbroso S, Sultan C. Transcriptional interferences between normal or mutant androgen receptors and the activator protein 1--dissection of the androgen receptor functional domains. Endocrinology. 1999;140:350–357. doi: 10.1210/endo.140.1.6418. [DOI] [PubMed] [Google Scholar]
- Lopez R, Garrido E, Pina P, Hidalgo A, Lazos M, Ochoa R, Salcedo M. HOXB homeobox gene expression in cervical carcinoma. Int. J. Gynecol. Cancer. 2006;16:329–335. doi: 10.1111/j.1525-1438.2006.00350.x. [DOI] [PubMed] [Google Scholar]
- Ma H, Hong H, Huang SM, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Miao J, Wang Z, Provencher H, Muir B, Dahiya S, Carney E, Leong CO, Sgroi DC, Orsulic S. HOXB13 promotes ovarian cancer progression. Proc. Natl. Acad. Sci. U S A. 2007;104:17093–17098. doi: 10.1073/pnas.0707938104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JD, Joseph JD, Sherk AB, Juzumiene D, Turnbull PS, Rafferty SW, Cui H, Anderson E, Fan D, Dye DA, et al. Differential presentation of protein interaction surfaces on the androgen receptor defines the pharmacological actions of bound ligands. Chem. Biol. 2009;16:452–460. doi: 10.1016/j.chembiol.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley BW. Coregulators: from whence came these "master genes". Mol. Endocrinol. 2007;21:1009–1013. doi: 10.1210/me.2007-0012. [DOI] [PubMed] [Google Scholar]
- Peterson TJ, Karmakar S, Pace MC, Gao T, Smith CL. The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor alpha transcriptional activity. Mol. Cell. Biol. 2007;27:5933–5948. doi: 10.1128/MCB.00237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherk AB, Frigo DE, Schnackenberg CG, Bray JD, Laping NJ, Trizna W, Hammond M, Patterson JR, Thompson SK, Kazmin D, et al. Development of a small-molecule serum- and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Res. 2008;68:7475–7483. doi: 10.1158/0008-5472.CAN-08-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ueno N, Kuroiwa A. Hox proteins functionally cooperate with the GC box-binding protein system through distinct domains. J. Biol. Chem. 2003;278:30148–30156. doi: 10.1074/jbc.M303932200. [DOI] [PubMed] [Google Scholar]
- Swinnen JV, Esquenet M, Goossens K, Heyns W, Verhoeven G. Androgens stimulate fatty acid synthase in the human prostate cancer cell line LNCaP. Cancer Res. 1997;57:1086–1090. [PubMed] [Google Scholar]
- Swinnen JV, Van Veldhoven PP, Esquenet M, Heyns W, Verhoeven G. Androgens markedly stimulate the accumulation of neutral lipids in the human prostatic adenocarcinoma cell line LNCaP. Endocrinology. 1996;137:4468–4474. doi: 10.1210/endo.137.10.8828509. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Tazawa S, Yawei Z, Katayama H, Kato Y, Nishiwaki K, Yokohama Y, Ishikawa M. Suppression of invasive characteristics by antisense introduction of overexpressed HOX genes in ovarian cancer cells. Int. J. Oncol. 2006;28:931–938. [PubMed] [Google Scholar]
- Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. U S A. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeltser L, Desplan C, Heintz N. Hoxb-13: a new Hox gene in a distant region of the HOXB cluster maintains colinearity. Development. 1996;122:2475–2484. doi: 10.1242/dev.122.8.2475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.