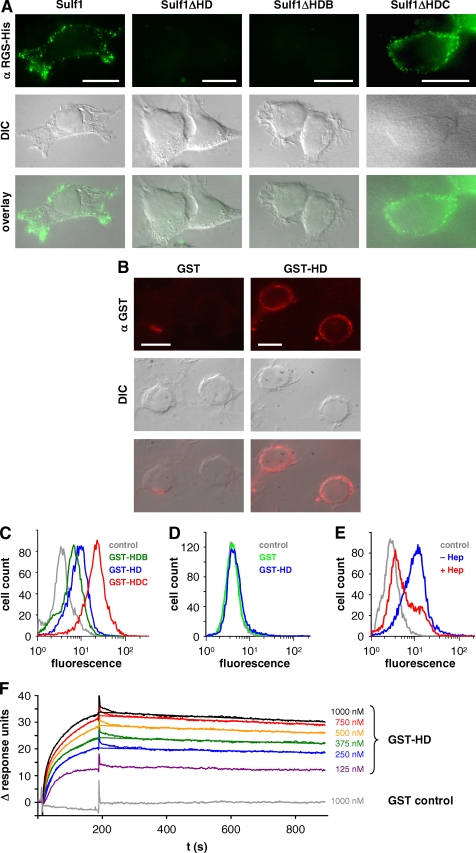
FIGURE 7.
Cell surface targeting of Sulf1 and binding of the HD to heparan sulfate and heparin. Non-permeabilized cells from HT1080 cell lines expressing full-length Sulf1, Sulf1ΔHD, -ΔHDB, or -ΔHDC were analyzed by indirect immunofluorescence microscopy with an antibody directed against the C-terminal RGS-His6 tag (A). Bars represent 10 μm. The overlays of the respective fluorescence images with differential interference contrast (DIC) images reveal a cell surface localization of Sulf1 and Sulf1ΔHDC. As a complementary approach to the deletion mutants, purified GST-HD was added to untransfected HT1080 cells and detected on the cell surface using anti-GST antibodies (B). GST was used as a negative control. The binding of the different fusion proteins GST-HD, GST-HDB, and GST-HDC was compared by FACS analysis (C). In the case of panel D, the HT1080 cells were pre-treated with trypsin to remove all cell surface proteins and extensively washed before GST-HD was applied. The experiment was repeated with cells that had instead been incubated with heparinases to remove HS chains of cell surface HSPGs (E). Binding of GST-HD to immobilized heparin was further analyzed by surface plasmon resonance studies as described in the text and compared with GST as a control (F). The data from 250 to 1000 nm GST-Sufl1HD were used for curve fitting, based on the heterogeneous ligand model as described in the text (χ2 = 0.046).