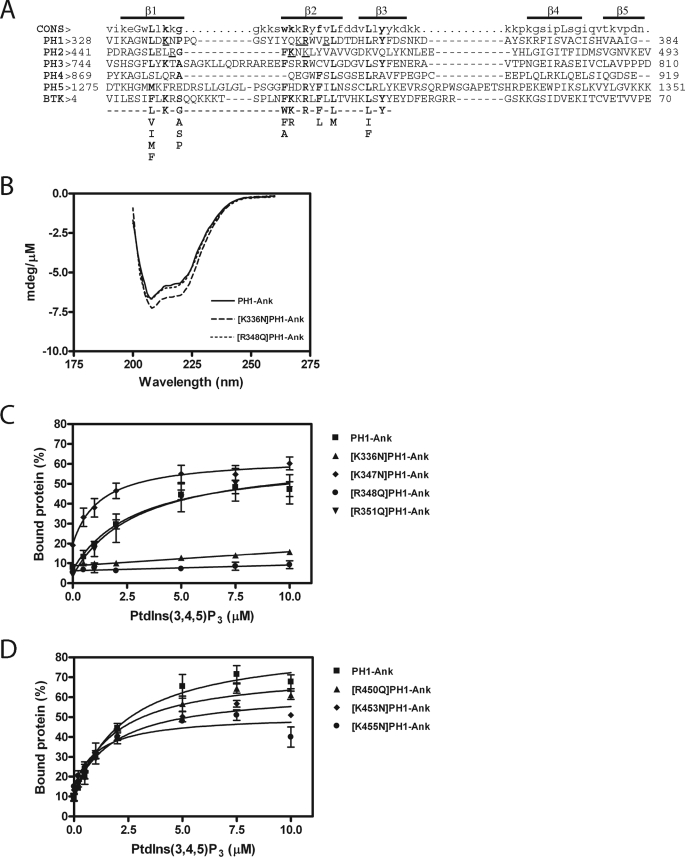
FIGURE 2.
A, alignment of ARAP1 PH domains with the PH domain of the human Bruton's tyrosine kinase (GenBankTM accession number Q06187) and the consensus sequence for PtdIns(3,4,5)P3 binding PH domain-containing protein. Residues critical for binding to PtdIns(3,4,5)P3 are shown in bold type (33, 34). Residues that have been mutated in the first and second PH domains and tested in this study are underlined. The regions predicted to form β1, β2, β3, β4, and β5 strands are indicated. B, circular dichroism spectra of [K336N]PH1-Ank and [R348Q]PH1-Ank. CD spectra were determined for the indicated recombinant protein as described under ”Experimental Procedures.“ C and D, effect of mutations in PH1 (C) and PH2 (D) domains on PtdIns(3,4,5)P3-dependent binding. 1 μm PH1-Ank or mutant PH1-Ank was incubated with sucrose-loaded LUVs (total phospholipid concentration of 1 mm) formed by extrusion through a 1-μm pore size filter. LUVs contained the indicated concentrations of PtdIns(3,4,5)P3. The concentration of phosphatidylinositol was adjusted in each LUVs to maintain a total phosphoinositide concentration of 50 μm. Vesicles were precipitated by ultracentrifugation, and associated proteins were separated by SDS-PAGE. The amount of precipitated protein was determined by densitometry of the Coomassie Blue-stained gels with standards on each gel. The results presented are the average ± S.E. of at least three experiments.