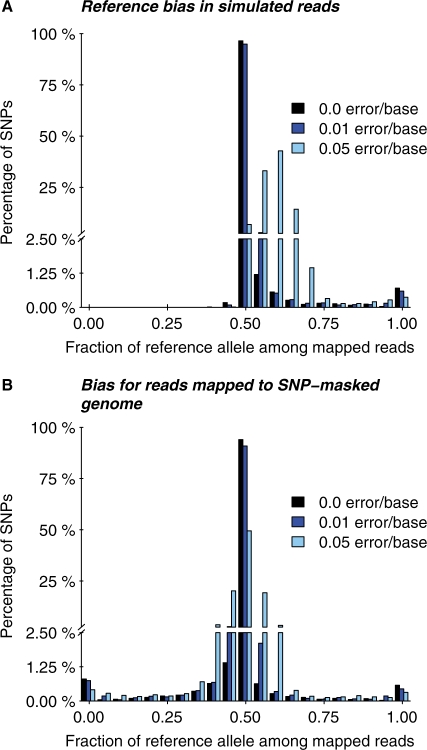
Fig. 2.
Magnitude of read-mapping biases in simulated data. (A) The distribution (across SNPs) of the proportion of correctly mapped reads that carry the reference allele, compared with the non-reference allele. The y-axis is broken into two segments to show more clearly the rates of highly biased SNPs. Three different rates of sequencing errors are shown. (B) Read-mapping was performed as in (A), except that the reads were aligned against a version of the genome sequence in which all SNP locations were masked. Notice that for both analysis methods, some SNPs are strongly biased, and that SNP masking does not clearly improve the results. Sequencing errors can substantially increase the extent of bias.