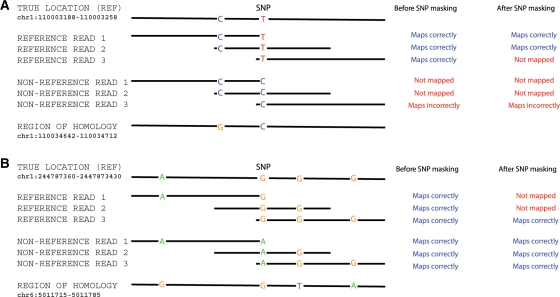
Fig. 3.
Two examples in which homology with other genomic locations leads to read-mapping biases. (A) Example of a SNP where there is a bias toward the reference allele before and after SNP masking (rs506008) and (B) example of a SNP where there is a bias toward the non-reference allele after SNP masking (rs11585481). Each example shows the variable sites in: (top row) the reference version of the genome sequence in the true location; (next six rows) three sample reads carrying the reference and three sample reads carrying the non-reference alleles at the SNP and (bottom row) the sequence in a region of homology elsewhere in the genome. The right-hand columns show how each read is mapped with, and without SNP masking. In these examples a read is mapped to a particular location if it has a unique best match at that location, and is unmapped if there is a tie between possible locations. The SNP masking generates an 1 nt mismatch between both alleles and the reference sequence at the masked site.