Fig. 2.
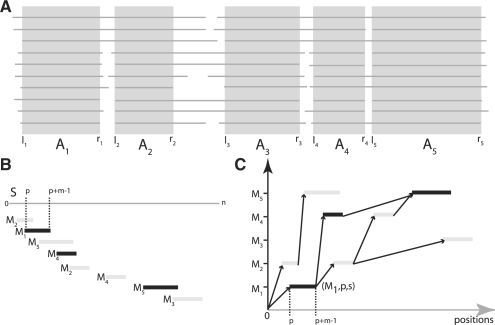
(A) Non-overlapping alignment blocks, excised from ungapped regions of a multiple alignment. Since li≤ri<lj≤rj for 1≤i≤j≤5, A=A1, A2, A3, A4, A5 is an ordered sequence of non-overlapping alignment blocks suitable to construct a PSSM-FM ℳ=M1, M2, M3, M4, M5. (B) Matches of Mi, i∈[1, 5], on sequence S, sorted in ascending order of their start position. (C) Graph-based representation of the matches of Mi, i ∈ [1, 5]. An optimal chain of collinear non-overlapping matches is determined, by computing an optimal path in the directed, acyclic graph. Observe that not all edges in the graph are shown in this example and that the optimal chain (indicated here by their black marked members) is not necessarily the longest possible chain.