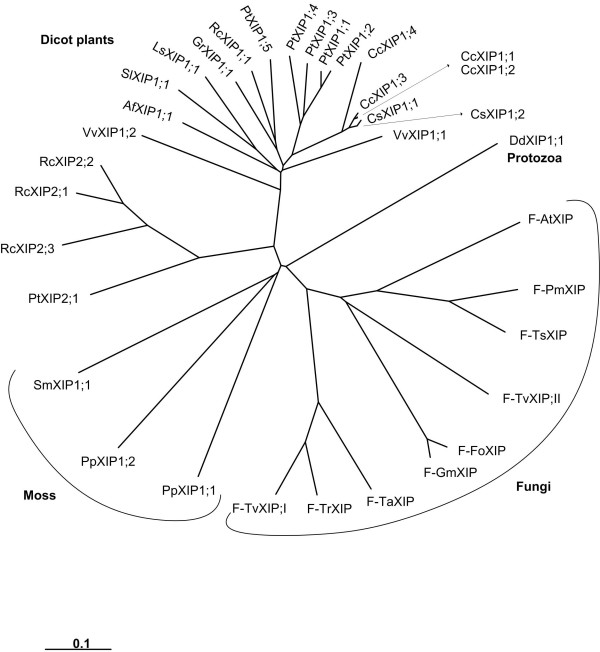
Figure 2.
Phylogenetic analysis of XIPs. All 35 XIPs from dicot plants, fungi, moss and protozoa have been used to construct the phylogenetic tree using NJ method. Multiple sequence alignment for creating the phylogenetic tree was generated by T-COFFEE. XIPs from dicot plants, fungi and moss cluster separately. All dicot XIPs cluster into two subgroups XIP1 and XIP2. All fungi XIPs and some dicot XIPs (Table 2) have been identified in this study. Other dicot XIPs, moss XIPs and the lone XIP from protozoa were identified by Danielson and Johanson. The names of Populus XIPs, dicot XIPs identified in this study and fungi XIPs are given in Tables 1, 2 and 3 respectively. The names of other dicot and the protozoan XIPs are given as follows with their IDs (EST/RefSeq/whole genome shotgun sequence) in brackets: SlXIP1;1 -- Solanum lycopersicum (BT014197), CcXIP1;1 -- Citrus clementina (DY275505), GrXIP1;1 -- Gossypium raimondi (CO092422), RcXIP1;1 -- Ricinus communis (EG656577), RcXIP2;1 -- Ricinus communis (EG666650), AfXIP1;1 -- Aquilegia formosa × Aquilegia pubescens (DR936893 and DT742029), DdXIP1;1 -- Dictyostelium discoideum (XM_639170) and VvXIP1;1 and VvXIP1;2 -- Vitis vinifera (AM455454).