Abstract
The roles in brain development. Previous studies have shown the association between OTX2 and OTX1 with anaplastic and desmoplastic medulloblastomas, respectively. Here, we investigated OTX1 and OTX2 expression in Non-Hodgkin Lymphoma (NHL) and multiple myeloma. A combination of semiquantitative RT-PCR, Western blot, and immunohistochemical analyses was used to measure OTX1 and OTX2 levels in normal lymphoid tissues and in 184 tumor specimens representative of various forms of NHL and multiple myeloma. OTX1 expression was activated in 94% of diffuse large B-cell lymphomas, in all Burkitt lymphomas, and in 90% of high-grade follicular lymphomas. OTX1 was undetectable in precursor-B lymphoblastic lymphoma, chronic lymphocytic leukemia, and in most marginal zone and mantle cell lymphomas and multiple myeloma. OTX2 was undetectable in all analyzed malignancies. Analysis of OTX1 expression in normal lymphoid tissues identified a subset of resting germinal center (GC) B cells lacking PAX5 and BCL6 and expressing cytoplasmic IgG and syndecan. About 50% of OTX1+ GC B cells co-expressed CD10 and CD20. This study identifies OTX1 as a molecular marker for high-grade GC-derived NHL and suggests an involvement of this transcription factor in B-cell lymphomagenesis. Furthermore, OTX1 expression in a subset of normal GC B cells carrying plasma cell markers suggests its possible contribution to terminal B-cell differentiation.
Fn1Growing evidence indicates that molecular mechanisms controlling cell-growth, differentiation, and cell-death are frequently recruited in different body organs and operate during embryonic development and postnatal life. Abnormal functioning of these mechanisms is frequently associated with or responsible for multiple diseases including cancer.1,2 This has suggested that mispatterning and/or abnormal positional information may be functionally involved in the initiation and/or maintenance of tumorigenesis. Several signaling pathways (eg, the SHH, WNT, and BMP pathways) and transcription factors (eg, GLI, RB, PAX, and HOX genes) have been implicated in various cancers.3,4,5,6 OTX1 and OTX2 are transcription factors containing a bicoid-like homeodomain and represent the vertebrate homologoues of the Drosophila orthodenticle gene. In mice, OTX1 and OTX2 genes are required for specification, maintenance, and patterning of forebrain and midbrain as well as for neuronal differentiation.7,8,9 Both genes are also required in the visual and acoustic sense organ development and OTX1 for corticogenesis, transient control of pituitary levels of GH, FSH, and LH hormones.7,9 In the hematopoietic system, OTX1 is required for the development of the erythroid compartment.10 Recently, it has been reported that, in humans, the OTX2 gene is amplified in a relevant percentage (20%) of primary anaplastic medulloblastomas and expressed at high levels in most of them, suggesting that it may represent a medulloblastoma oncogene.11,12 Similarly, OTX1 is overexpressed in medulloblastomas of the nodular/desmoplastic subtype.13 OTX1 and OTX2 expression was not detected in other brain tumors including astrocytomas, glioblastomas, oligodendrogliomas, meningiomas, ependymomas, or in several tumors of non-neural origin affecting breast, thyroid, prostate, liver, lung, stomach, pancreas, kidney, and colon (data not shown).11,12,13 Here, we investigated the expression of OTX1 and OTX2 in B-cell Non-Hodgkin Lymphoma (NHL). These tumors represent an heterogeneous group of malignancies arising from mature B-cells recruited in germinal centers (GCs) of secondary lymphoid organs during a T-cell dependent immune response.14,15 Our results demonstrate that OTX1 but not OTX2, is constitutively expressed in specific subsets of B-cell NHL. The concurrent analysis of a panel of nonmalignant cells and tissues of the hematopoietic system indicates that OTX1 is expressed in a minor subset of GC-restricted B-cells displaying a plasma cell phenotype. Altogether, these findings identify constitutive expression of OTX1 in NHL subtypes as a transformation-associated event, while its presence in a restricted subset of non-transformed GC B-cells suggests a potential involvement in plasma cell differentiation.
Materials and Methods
Lymphoma Tissue Samples
For all lymphoma cases investigated, both paraffin-embedded and fresh tumor samples at diagnosis were available. Cases were retrieved from tissues and nucleic acid banks of the Pathology and Hematology-Oncology Units of the National Cancer Institute of Naples, Fondazione Pascale. According to local institutional guidelines, all patients provided informed consent to use biological material obtained during diagnostic procedures for preclinical investigations. In addition, the Scientific Review Board of the Istituto Nazionale Tumori, Fondazione G. Pascale, IRCCS has approved the study here presented (protocol DSC/2104). In selected cases, tissue samples were obtained on biopsy of other lymphoma-involved tissues, including mediastinal masses, rhinopharynx, gastric mucosa, testis, and spleen. In the case of Multiple Myeloma (MM) and B-cell Small Lymphocytic Lymphoma/Chronic Lymphocytic Leukemia (B-SLL/CLL), anticoagulated bone marrow (BM) aspirates and peripheral blood (PB) samples with more than 80% tumor cells were used for RNA extraction. In selected MM cases, tumor plasma cells were further processed to >95% purity by magnetic immunoselection with anti-CD138 antibodies and MiniMACS columns (Miltenyi Biotec; Calderara di Reno (Bologna), Italy).16 The lymphoma cases were classified according to the current World Health Organization classification17 and characterized by immunophenotypic studies (TdT, CD79a, CD20, CD23, CD5, CD3, CD56, CD43, CD30, CD34, CD15, CD45, EMA, Cyclin D1, and Ki-67). Diffuse Large B-cell Lymphoma (DLBCL) were further classified into GC-like and activated B-cell-like subsets by means of CD10, BCL-2, BCL-6, MUM-1, and CD138 immunostainings.18,19 Diagnoses were integrated by detection of the t(14;18)(q32;q21), t(11;14)(q13;q32), and t(11;18)(q21;q21) translocations for follicular lymphoma, mantle cell lymphoma, and extranodal marginal zone B-cell lymphomas of the mucosa-associated lymphoid tissue, respectively.
Nonmalignant Lymphoid Tissue Samples and Isolation of Normal Lymphoid Cells
Nonmalignant lymph nodes were obtained during surgical procedures for solid tumors and checked for the absence of tumor cells by histopathology. Reactive lymph nodes were obtained from patients with a final histopathological and molecular (ie, absence of clonal VDJ rearrangements) diagnosis of reactive follicular hyperplasia. Nonmalignant tonsil tissues were obtained from adult patients undergoing tonsillectomy. Mononuclear cells from PB of healthy donors (n = 10) and from single-cell suspension of tonsil tissues (n = 4) were isolated by centrifugation on a Ficoll-Hypaque gradient and further purified by sequential direct immunomagnetic selection (Miltenyi system) over MiniMACS columns in the following cell fractions: CD3+ (T cells); CD19+ (B cells); and CD3−/CD19−(non-T cells and non-B cells).16
Isolation of GC B-Cell Subsets
The purification of GC centroblasts (CBs) and centrocytes (CCs) was based on differential CD77 expression by using the MidiMACS magnetic sorting system (Miltenyi Biotec), as previously described.20 Staining for the GC-specific marker BCL-6 confirmed the expression of the transcriptional repressor in over 80% of purified CBs and CCs.
RT-PCR Analysis for OTX1, OTX2, BCL6, and PAX5
Total RNA was extracted with Trizol and treated with superscript II Rnase H− reverse transcriptase. For RT-PCR analysis, calibration experiments were performed to determine optimal nonsaturating PCR conditions for each pair of primers. We tested amplification at 25, 27, 29, and 31 cycles and found best conditions at 29 cycles for OTX1, 27 for BCL6 and PAX5, and 23 cycles for β-ACTIN. As for OTX2, we used saturating conditions (32 cycles) to detect also low levels of mRNA. A human medulloblastoma cDNA sample was used as a positive control in RT-PCR experiments.
Primer sequences and length of amplified products are as follows (forward and reverse primers), respectively: OTX1 (5′-AGACGCATCAGACCCTGAAGGACT-3′, 5′-CCAGACCTGGACTCTAGACTC-3′ 355 bp); OTX2 (5′-GTACCCCCGATTTGGGCCGACT-3′, 5′-ATCTAGCTGCGCCCGAGTGAAC3′-198 bp); PAX5 (5′-CCAGAGGATAGTGGAACTTGCTC-3′, 5′-GCTAGGCACGGTGTCATTGTCAC-3′ 280 bp); BCL6 (5′-GAGCATGTTGTGGACACTTGCCG-3′, 5′-GACATCCCGAAACTCCTCATCG-3′ 296 bp); and β-ACTIN (5′-CCAGGTCATCACCATTGGCAATG-3′, 5′-GCTGATCCACATCTGCTGGAAGGT-3′, 340 bp).
Structural Analysis of OTX1 Gene in Malignant Cells
To assess the integrity of the OTX1 coding sequence, we amplified the gene in two independent reactions spanning together the entire coding region plus flankings and overlapping by 112 bp. The 5′ region (from 82 bp upstream of methionine to aa 207) was amplified by using the following primers: forward primer, 5′-AGACGCATCAGACCCTGAAGGACT-3′, reverse primer, 5′-CTGCATACACGAGGTGTTGCTAGG-3′. The 3′ region (from aa 171 to 39 bp downstream of the stop codon) was amplified by using the following primers: forward primer, 5′-ACCAGCTGCCTCATCTATCTGGAG-3′, reverse primer, 5′-GCAGGTAGTTGCGTTTCTTCTCCTC-3′. A subcloned OTX1 full-length cDNA fragment was used as template to generate PCR fragments of control size.
To assess abnormality in OTX1 gene copy number, we analyzed by real time-quantitative PCR DNAs extracted from 15 cases of DLBCL and 8 cases of SLL/CLL. Real time-quantitative PCRs to assess the OTX1 gene copy number were performed on the ABI/Prism 7700 sequence detector (Applied Biosystems, Monza, Italy) platform, by the Sybr Green I dye methodology. For OTX1, forward primer: 5′-TAACCCTACGCCCTCCTCTTCCTACT-3′; reverse primer: 5′-AAGCAGTCGGCAGAGTTGAAGGCAAG-3′. To normalize the DNA amount, a simultaneous RT-quantitative PCR for the ALBUMIN gene was run in duplicate. For ALBUMIN, forward primer: 5′-TGAAACATACGTTCCCAAAGAGCCT-3′; reverse primer: 5′-CTCTCCTTCTCAGAAAGTGTGC-3′. For quantitation, the ratio between the cycle threshold (CT) of OTX1 and the CT of ALBUMIN from each tumor sample was compared with the ratio between the CT of OTX1 and the CT of the ALBUMIN derived from the analysis of the DNA calibrator.
Immunohistochemistry, Western Blot Analysis, and Cell Counting of OTX1+ Cells
The mouse primary antibodies and dilution used are as follows: anti-PAX5 (BD, Biosciences Pharmingen San Diego, CA) 1:100; anti-CD20 (Dako Glostrup, Denmark) 1:400; anti-CD34 (Dako) 1:50; anti-BCL6 (Dako) 1:50; anti-CD68 (Abcam, Cambridge, UK) 1:300; anti-CD7 (Dako) 1:25; anti-CD45 (Dako) 1:100; anti-CD138 (Dako) 1:300; anti-CD3 (Dako) 1:200; anti-CD10 (Serotec, Martinsried, Germany) 1:80; anti-IgG (Serotec) 1:200; anti-IgM (Dako) 1:200; anti-IgE (Dako) 1:200; anti-p27kip1 (BD Pharmingen) 1:1500; anti-Ki-67 (Dako) 1:200; anti-Ph-H3 (Sigma, St. Louis, MC) 1:2000; and anti-CycA (Chemicon) 1:200. The rabbit anti-OTX was kindly provided by G. Corte and used at a dilution of 1:3500. For immunohistochemistry on wax-embedded tissues, antigens were unmasked by four rounds of microwave boiling (4 minutes/boiling) at 700 Watts in sodium citrate buffer (10 mMol/L pH 6)20 and primary antibodies were applied to the sections and left overnight at room temperature. After washing in PBS, secondary antibodies were applied for 60 minutes, then the slides were PBS washed and processed for their observation and image capture.
For immunohistochemistry, primary antibodies were revealed by carbazole staining of horseradish peroxidase-coniugated secondary antibodies by using the Dako Envision plus kit following the manufacturer’s instructions. For immunofluorescence, primary antibodies were revealed by using anti-mouse or anti-rabbit or anti-goat Alexa-594 or Alexa-488 or Alexa-350 secondary antibodies.
For Western blot analysis, protein extracts from nonmalignant lymph nodes, DLBCL lymph nodes, and E12.5 mouse embryos as a control, were obtained by homogenizing the specimens in a lysis buffer containing a cocktail of protease inhibitors. The supernatants were probed in a standard Western blot with the anti-OTX polyclonal antibody (1:10000), and revealed by Enhanced Chemiluminescence.
The counting of OTX1+ cells in the GCs of three tonsils and four nonmalignant Lymph Nodes was performed by analyzing adjacent sequential sections (9-μm thick) covering 50 GCs for each sample. For each GC, the section containing the highest number of OTX1+ cells was taken into consideration. Sections were captured, printed, and OTX1+ cells were counted. GCs considered in this analysis were equally selected in periphery and more central area of the tissue specimens. Based on the number of OTX1+ cells, we arbitrarily defined two major groups of GCs: the first containing from 1 to 4 OTX1+ cells; and the second from 5 to the highest number of OTX1+ cells scored (n = 65). This second group was further analyzed to define the most represented subgroup, which in tonsil and nmLN contains between 8 and 20 OTX1+ cells.
Results
Expression of OTX1 and OTX2 in NHL
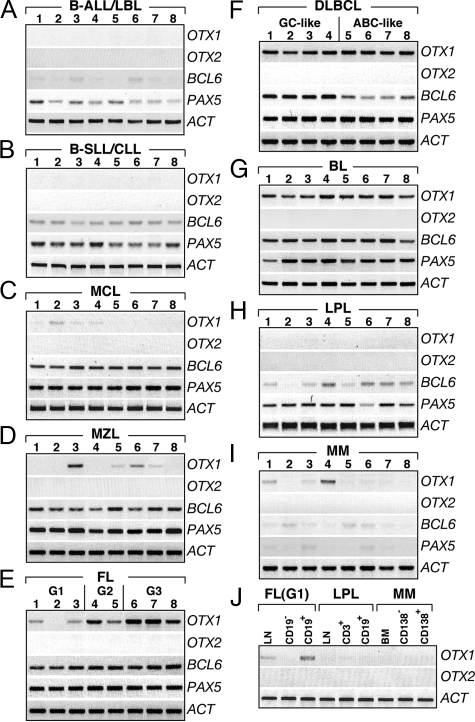
To study the relative level of OTX1 and OTX2 mRNAs in B-cell lymphoma, a total of 184 pathological samples representative of different lymphoma subtypes were analyzed by RT-PCR (Table 1). These included precursor B-Acute Lymphoblastic Lymphoma/Leukemia (B-ALL/ B-LBL), B-SLL/CLL, Mantle Cell Lymphoma (MCL), Marginal Zone Lymphoma (MZL), Follicular Lymphoma (FL), DLBCL, Burkitt lymphoma (BL), Lymphoplasmacytic Lymphoma (LPL) and MM (Materials and Methods). OTX1 was not detected in B-ALL/B-LBL (n = 11), B-SLL/CLL (n = 25), and LPL (n = 10) cases (Figure 1A, B, and H; Table 1), while only 3 out of 14 MCL showed low level of OTX1 expression (Figure 1C). In MZL (n = 16) (Figure 1D, Table 1), OTX1 was detected at low level in three cases, at a moderate level in one case, while it was undetectable in the remaining 12 cases.
Table 1.
OTX1 Expression in B-Cell Non-Hodgkin Lymphoma Subtypes
| Lymphoma subtype | Total positive* | Percent positive |
|---|---|---|
| B-ALL/B-LBL | 0/11 | 0 |
| SLL/CLL | 0/25 | 0 |
| MCL | 3/14 | 21 |
| MZL | 4/16 | 25 |
| Extranodal | 1/6 | 17 |
| Nodal | 3/7 | 43 |
| Splenic | 0/3 | 0 |
| FL | 25/41 | 61 |
| Grade 1 | 8/17 | 46 |
| Grade 2 | 7/13 | 53 |
| Grade 3† | 10/11‡ | 90 |
| DLBCL | 31/33 | 94 |
| BL | 10/10 | 100 |
| LPL | 0/10 | 0 |
| MM | 3/24 | 12 |
As detected by RT-PCR (Materials and Methods).
Including four cases of FL G3A and seven cases of FL G3B.
The negative sample is from a patient with FL G3A.
Figure 1.
OTX1 and OTX2 expression in B-cell lymphomas. OTX1 and OTX2 expression analysis in representative pathological samples of B-ALL/LBL (A), B-SLL/CLL (B), MCL (C), MZL (D), FL (E), DLBCL (F), BL (G), LPL (H), and MM (I) shows that OTX2 is not transcribed in any of the pathological samples, whereas OTX1 exhibits high level of expression in all of the patients with FL G3 (E), DLBCL (F), and BL (G). Moderate OTX1 expression is detected in FL G2 (E) and low level of OTX1 is found in most of the FL G1 (E). OTX1 is sporadically expressed at low level in patients with MCL (C), MZL (D), and MM (I); no expression is detected in B-ALL/LBL (A), B-SLL/CLL (B), and LPL (H). High level of PAX5 and BCL6 expression is consistently detected in MCL (C), MZL (D), FL (E), GC like DLBCL (F), and BL (G) samples. J: OTX1 expression in LN, CD19− and CD19+ cells isolated from a representative FL (G1); LN, CD3+ and CD19+ cells isolated from a LPL; and BM, CD138− and CD138+ cells isolated from a MM. G1, G2 and G3 indicate the histological grade of the FL samples. LN tissues are analyzed in all cases, except for lanes 4 to 8 of B where PB tumor cells are used; all lanes of I correspond to BM; lanes 1, 2 of D correspond to splenic tissue and lanes 7, 8 of D correspond to extranodal tissue from gastric biopsies. RT-PCRs are performed at 29 cycles for OTX1, 32 cycles for OTX2, 27 cycles for PAX5 and BCL6, and 23 cycles for β-ACTIN.
OTX1 was highly expressed in the majority of FL and, virtually, in all of the DLBCL and BL cases analyzed. Specifically, in FL OTX1 transcripts were detected in 25 out of 41 cases (Table 1). Interestingly, all grade-3 (G3) FL exhibited high level of OTX1 transcripts while those belonging to the G2 and G1 types showed respectively moderate and low expression (Figure 1E). Moreover, 53% of FL G1 and 46% of FL G2 cases did not express OTX1 as opposed to only 10% of FL G3 cases (Table 1). OTX1 expression in three FL of G1 type was confirmed in CD19+ purified B-cells (Figure 1J and data not shown). As predicted, the transcription factors PAX5 and BCL6 were uniformly expressed in all FL (Figure 1E). OTX1 was homogeneously expressed at high level in roughly all DLBCL and BL cases tested (Figure 1, F and G; Table 1). In DLBCL, OTX1 was found highly expressed in both the GC- (n = 14) and the activated B-cell (n = 17) types. Finally, only 3 out of 24 MM cases expressed low level of OTX1 (Figure 1, I and J). Absence of OTX1 expression was also confirmed by analyzing CD138+ tumor plasma cells purified from the BM of five MMs (Figure 1J). Similarly, the absence of OTX1 expression in LPL cases was confirmed by analyzing the CD19+ tumor cell population (Figure 1J). In contrast to a positive control represented by a human medulloblastoma sample, OTX2 expression was never detected in any of the B-cell lymphoma subtypes analyzed (Figure 1, A–J, and data not shown). Altogether, these data indicate that OTX1 expression is preferentially activated in GC B-cell-derived NHL subtypes, representing a novel molecular marker to identify FL G3, DLBCL, and BL cases.
Activation of OTX1 Expression in DLBCL and BL is not Linked to Genomic Rearrangements and/or Gene Amplification
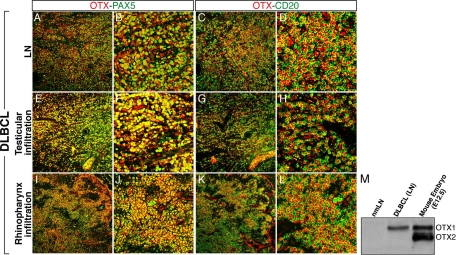
The distribution and levels of the OTX1 protein were analyzed in tumor tissues from five representative DLBCL cases by using an anti-OTX antibody recognizing both OTX1 and OTX2.21 All of the five DLBCL cases exhibited a uniform distribution of OTX1 in the nucleus of the PAX5+-CD20+ tumor cells (Figure 2, A–D, and data not shown) and, compared with nmLNs, a remarkably high protein level (Figure 2M and data not shown). In one case, homogenous strong high level of OTX1 protein was confirmed in testicular and rinopharynx infiltrations (Figure 2, E–L). Since in humans the OTX1 gene maps to chromosome 2 p13-15,22 a region frequently rearranged in DLBCL23 and given the frequent OTX2 gene amplification in primary medulloblastomas, we analyzed the OTX1 gene dosage in 15 DLBCL and the integrity of its coding region in 13 DLBCL and 7 BL. No alterations were detected in both copy number and structure of the OTX1 coding sequence (data not shown), ruling out the possibility that deregulated OTX1 expression in NHL resulted from recurrent genomic aberrations at the OTX1 gene locus.
Figure 2.
Immunodetection of OTX1 in DLBCL tissues. Co-immunohistochemistry experiments with OTX and PAX5 (A, B, E, F, I, J) and OTX and CD20 (C, D, G, H, K, L) antibodies in the LN (A–D), testicular (E–H), and rhinopharynx (I–L) infiltrations of a DLBCL show robust expression and full co-localization of OTX1 with both PAX5 and CD20 in tumor cells. M: Western blot analysis showing the OTX1 protein level in extracts from nmLN, DLBCL LN, and mouse embryos at gestational day 12.5 (E12.5). B, F, and J, and D, H, and L are magnifications corresponding to a restricted area of A, E, and I, and C, G, and K, respectively; the mouse and human OTX1 proteins differ by a single amino acid.
Expression of OTX1 and OTX2 in Nonmalignant Lymphoid Cells and Tissues
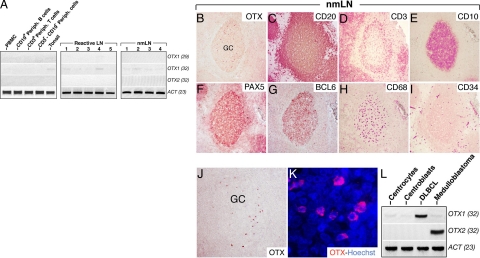
Given the strong expression of OTX1 in GC-derived B-cell malignancies, we studied its expression and that of OTX2 in the normal B-cell counterpart. RNAs from tonsils (n = 4), total peripheral mononuclear cells (n = 6), CD19+ B-cells (n = 5), CD3+ T cells (n = 4), and CD19−/CD3− double negative mononuclear cells (n = 2) purified from PB of healthy donors were analyzed by RT-PCR for OTX1 and OTX2. None of the normal lymphoid cell populations tested exhibited detectable levels of OTX1 mRNA, except for a weak expression in unfractionated tonsillar cells at a number of PCR cycles (n = 32) remarkably higher than that used for the study of the pathological tissue samples (n = 29) (Figure 3A and data not shown). A similar low level of OTX1 expression was observed in reactive and nmLNs (Figure 3A). OTX2 transcripts were never detected in the same samples (Figure 3A). These findings may either reflect high OTX1 expression in a limited number of nmLN and tonsil cells or low OTX1 level expressed by the majority of the resident cells. Furthermore, expression data on reactive tonsils rule out the possibility that high OTX1 level found in tumor samples depends exclusively on the proliferation rate of the transformed cells.
Figure 3.
OTX1 expression in normal lymphoid cell populations, reactive LNs, and nmLNs. A: OTX1 expression analysis in peripheral blood mononuclear cells (PBMC), CD19+ peripheral B-cells, CD3+ peripheral T-cells, CD3−/CD19− peripheral lymphocytes, total tonsil tissue, five LNs with reactive follicular hyperplasia (reactive LN), and four nmLNs shows a faint signal only at 32 PCR cycles in tonsil, reactive LNs, and nmLNs, while no signal is detected for OTX2 in all of the samples. Immunodetection of OTX1 (B and J), CD20 (C), CD3 (D), CD10 (E), PAX5 (F), BCL6 (G), CD68 (H), and CD34 (I) by using carbazole staining in adjacent sections of a nmLN shows that, according to the gross distribution of GC cell-markers, a few OTX1+ cells are detected only in the GC area. K: OTX immunohistochemistry in Hoechst-stained GC cells shows that OTX1 is prevalently localized to the cytoplasm. L: OTX1 and OTX2 RT-PCR show a very low level of OTX1 transcripts in both CCs and CBs while no expression for OTX2 is detected. As a positive control, the expression of OTX1 and OTX2 is shown in DLBCL and medulloblastoma, respectively. GC, germinal center.
OTX1 Expression Pattern in Normal Lymphoid Tissues
To assess the expression pattern of OTX1 at the single-cell level, immunohistochemical analysis on nmLN and tonsil tissues was performed.
OTX1+ cells were detected in low number, preferentially within the BCL6+ GC areas, in both nmLNs and tonsils (Figure 3B, G, J). Noteworthy, OTX1+ cells were not detected in all of the GCs scored in these tissues. Cell-counting performed in three tonsils and four nmLNs (Material and Methods) showed that respectively 88% (n = 132 of 150) and 74% (n = 148 of 200) of GCs contained between 5 and 65 OTX1+ cells. Among the scored GCs, 63% (n = 83 of 132) in tonsils and 71% (n = 105 of 148) in nmLNs displayed between 8 and 20 OTX1+ cells and defined the most representative subgroup. OTX1+ cells were not detected in the interfollicular areas and in the mantle zone (Figure 3, B and J, and data not shown) as confirmed from the histological analysis of serial sections stained with lineage-specific markers such as CD20, CD3, CD10, PAX5, BCL6, CD68, and CD34 (Figure 3, C–I). We also determined the subcellular localization of OTX1 in Hoechst-stained GC cells. Surprisingly, and in contrast with the nuclear-restricted distribution of OTX1 observed in NHL (Figure 2 and data not shown), in nmLNs the OTX1 protein appeared prevalently localized to the cytoplasm (Figure 3K). A similar intracellular distribution of the protein was observed in tonsils. Given the expression of OTX1 within the GC, we analyzed its expression in sorted populations of CCs and CBs. A comparable low level of OTX1 transcripts was detected in the two GC subsets (Figure 3L). In accordance with data on unfractionated tissues samples, expression of OTX2 was absent in the GC B-cell subsets (Figure 3L). These findings on normal lymphoid tissues indicate that OTX1 is (1) selectively expressed in a minor, distinct GC B-cell subset; and (2) predominantly localized to the cytoplasm of GC B-cells, in contrast to their transformed counterpart.
Phenotypic Characterization of OTX1+ B Cells within the GC
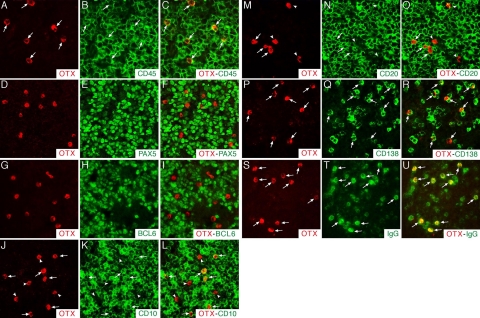
Next, we investigated at the single-cell level the phenotypic identity of OTX1+ cells in the GC of nmLN and tonsil sections. All of the OTX1+ cells expressed the pan-hematopoietic marker CD45 (arrows in Figure 4, A–C) and were CD34−, CD68−, CD7−, and CD3− (Supplemental Figure S1, C–J, see http://ajp.amjpathol.org), ruling out the possibility that they belong to the endothelial, macrophagic, histiocytic, or T-cell lineages. We also analyzed the expression of BCL6, which labeled most of the CBs and CCs, and the pan-B-cell marker PAX5. OTX1+ cells did not exhibit BCL6 nor PAX5 expression (Figure 4, D–I). Conversely, about 60% of the OTX1+ cells expressed CD10 and/or CD20 (arrows in Figure 4, J–O). Next, we analyzed the expression of the plasma cell marker CD138 and that of IgG, IgM, or IgE, which recognized the corresponding subsets of immunoglobulin-producing plasma cells. OTX1+ cells were CD138+ and expressed high levels of intracellular IgG (arrows in Figure 4, P–U), while none of them expressed IgM or IgE (data not shown). This expression analysis was confirmed at a low magnification (Supplemental Figure S1, see http://ajp.amjpathol.org). The unusual phenotype of OTX1+ cells was finally assessed by triple immunohistochemistry experiments. This experiment showed that all of the OTX1+ cells were CD138+ and about 30 to 40% of them co-expressed CD20 (Figure 5, A–C); the OTX1+CD20+ cells were consistently CD10+, while none of the OTX1+CD20− cells expressed CD10 (Figure 5, D–F); and none of the OTX1+ cells were PAX5+ (Figure 5, G–I). Finally, we determined the cell-cycle distribution of OTX1+ GC cells. Specifically, we measured the fraction of OTX1+ cells co-expressing cell-cycle associated markers such as Ki-67, p27kip1, CYCLIN A or phospho-histone 3. OTX1+ cells co-localized with the fraction of GC B-cells expressing p27kip1, while none of them expressed any proliferation marker (Supplemental Figure S2, see http://ajp.amjpathol.org and data not shown), thus indicating that the OTX1+ cells represent a quiescent cell population.
Figure 4.
Cell-identity analysis of OTX1+ cells in the GC of nmLN. Immunohistochemistry experiments to assess at single cell level co-localization between OTX1+ cells and those expressing CD45 (A–C), or PAX5 (D–F), or BCL6 (G–I), or CD10 (J–L), or CD20 (M–O), or CD138 (P–R), or IgG (S–U) show that OTX1+ cells never co-express PAX5 or BCL6, while several of them co-localize with CD10 and CD20, and all of them are CD45+, CD138+, and IgG+. The arrows point to OTX1+ cells co-expressing CD45, or CD10, or CD20, or CD138, or IgG; the arrowheads point to the cells expressing only OTX1.
Figure 5.
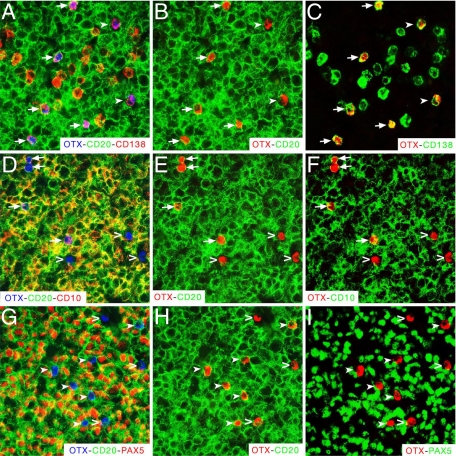
OTX1+ cells exhibited unusual plasma cell-like identity. Co-immunohistochemistry performed with OTX, CD20, and CD138 (A–C), OTX, CD20, and CD10 (D–F), and OTX, CD20, and PAX5 (G–I) antibodies on adjacent nmLN sections. In A–C the arrows point to the OTX1+-CD20+-CD138+ cells and the filled arrowheads to the OTX1+- CD20−-CD138+ cells; in D–F the arrows point to the OTX1+-CD20+-CD10+ cells and the open arrowheads to the OTX1+-CD20−-CD10− cells; in G–I the filled arrowheads point to the OTX1+-CD20+-PAX5− cells and the open arrowheads to the OTX1+-CD20−-PAX5− cells. The red color for OTX (B,C,E,F,H,I) and the green color for CD138 (C), CD10 (F), and PAX5 (I) are pseudocolors of the same immunohistochemistry shown in A, or D, or G.
Together these data suggest the existence of a novel quiescent subset of CD138+ GC B-cells marked by the expression of OTX1. An attractive possibility is that these postmitotic cells reflect a transient stage of GC B cell differentiation leading ultimately to the generation of antibody-secreting cells. Furthermore, based on the phenotypic characterization, OTX1+ cells found in normal GCs, likely do not correspond to those giving rise to DLBCL, BL, and FL malignancies.
Discussion
This study shows for the first time that the homeobox gene OTX1 is constitutively expressed in a subset of human B-cell malignancies. In particular, we have shown that OTX1 expression is preferentially restricted to tumors derived from GC and early post-GC B-cells. Indeed, pre- and post-GC B-cell malignancies including B-LL/B-ALL, B-SLL/CLL, and MM did not express OTX1 or exhibited a low expression in a small fraction of them (MCL and MZL) (Table 1). In contrast, OTX1 mRNA was detected in 61% of FL, in most DLBCL (94%), and in all cases of BL. In FL, OTX1 expression was associated to histological grading of tumors since G3-type tumors displayed the highest level of expression (90%) as compared with G2 (54%) and G1 (47%) cases.
Based on this expression profile, it appears that the molecular event(s) underlying OTX1 deregulation in human B-cell lymphomas is mostly restricted to precursor cells arising in the GC (FL, DLBCL of GC-like phenotype, and BL), or immediately poised to exit it (DLBCL of activated B-cell type).
Consistent with RNA expression data, immunolocalization experiments showed that in DLBCL tumor cells the OTX1 protein is detected in both primary and distal sites. This suggests that OTX1 induction might represent an early event occurring in the tumorigenic process. In lymphoma cells, the OTX1 protein exhibits the expected molecular weight and nuclear distribution. Our data indicate that OTX1 activation represents a molecular event specifically associated with mature B-cell lymphomagenesis. Indeed (1) purified CD19+, peripheral mononuclear CD3+ or CD19−CD3− non-B cells and non-T cells isolated from healthy donors lack detectable levels of OTX1 transcripts; (2) reactive follicular hyperplasia samples exhibit no increase in OTX1 expression, thus ruling out the possibility that OTX1 induction is a mere reflection of B-cell proliferation; and (3) in nmLN and tonsil OTX1 protein expression is detected in the cytoplasm of a subset of GC-restricted PAX5−-BCL6−-CD138+ cells. Collectively, these data suggest that the nonmalignant OTX1+ GC B cells do not represent the normal counterpart of the OTX1+ tumor cells; rather they support the possibility that activation of OTX1 expression is an independent molecular event occurring in FL, DLBCL, and BL tumor subtypes. The finding that the fraction of non-transformed OTX1+CD138+IgG+ GC B cells can be further subdivided in two major subgroups based on CD20 and CD10 expression, suggests the existence of distinct subsets of GC-restricted CD138+ cell populations. Interestingly, co-expression of OTX1 and CD138 was limited to a restricted subset of GC B cells, whereas it was detected neither in normal nor in transformed plasma cells present in extra GC sites. This result suggests that OTX1 expression may define a transitory stage in the developmental pathway of GC B cells leading ultimately to the generation of long-lived plasma cells.
An intriguing aspect of this study is the finding that in nonmalignant GC B-cells, OTX1 is prevalently localized to the cytoplasm, while in the lymphoma cells, the protein displays the predicted nuclear distribution. While the role of OTX1 in the cytoplasm of CD138+ GC cells remains to be deciphered, its induced expression in the nucleus of tumor cells may alter their gene expression profile and thus, determine abnormalities in cell-identity and/or proliferation.8,9 For OTX1 no direct proof of its involvement in cancer exists. However, similarly to OTX2, which is involved in the pathogenesis of anaplastic medulloblastoma, OTX1 expression has been correlated to the occurrence of nodular/desmoplastic medulloblastomas.13 This, together with the observation that in mouse OTX1 and OTX2 are functionally equivalent7,9 suggests that OTX1 could share with OTX2 an oncogenic activity in specific cell-context(s).
The OTX1 gene locus is located on human chromosome 2 at position p13-15,22 which, is frequently rearranged in mature B-cell lymphomas.23,24,25,26,27 Despite this, neither alterations in the OTX1 gene-copy number nor abnormalities in the length of its coding sequence were revealed.
In summary, this study provides new evidence indicating that OTX1 expression is activated in a large fraction of high-grade FL and in all DLBCL and BL where it is predominantly found in the nuclear compartment. These findings lead us to hypothesize a role for OTX1 in the onset and/or maintenance of GC-derived NHL. Also, the expression of OTX1 within normal GCs detected in a fraction of resting CD138+ IgG+ cells highlights the existence of subset of these cells with plasma cell commitment properties.
Supplementary Material
Acknowledgments
We thank Luigo Del Vecchio, Ulf Klein, and Tomasso Russo for critical reading of this article and helpful suggestions. We are grateful to Riccardo Dalla Favera and Ulf Klein for providing RNAs from purified centroblasts and centrocytes, and to Antonietta Secondulfo for article preparation.
D.O. and D.A. were responsible for performance of experiments and data analysis; F.R. collected and analyzed patient data; R.D.F. performed experiments and collected data; V.S. performed experiments; R.D.F. performed experiments; F.F. collected tissue samples; P.M. collected data; A.D.C. performed pathological diagnosis and provided tissue samples; S.C. analyzed and interpreted results, and drafted the article; A.P. designed research, analyzed data, and drafted the article; A.S. developed the study, designed research, analyzed data, performed experiments, and drafted the article.
Footnotes
Address reprint requests to Antonio Pinto, Hematology-Oncology Unit and Stem Cells Transplantation, Istituto Nazionale Tumori, Fondazione ‘G. Pascale’, I.R.C.C.S. Via M. Semmola, 80131 Naples, Italy. E-mail: apinto.int.napoli@tin.it; or Stefano Casola, IFOM-The FIRC Institute of Molecular Oncology, IFOM-IEO Campus, Via Adamello 16, 20139 Milan, Italy. E-mail: stefano.casola@ifom-ieo-campus.it; or Antonio Simeone, CEINGE Biotecnologie Avanzate, Via Comunale Margherita 482, 80145 Naples, Italy. E-mail: simeone@ceinge.unina.it.
Supported by the Italian Association for Cancer Research, the FP6 for the European Transcriptome, Regulome and Cellular Commitment Consortium (EuTRACC) Integrate Project (LSHG-CT-2007-037445), and the Fondazione Cassa di Risparmio di Roma (to A.S.), the Ministero della Salute, Ricerca Finalizzata, Fondo Sanitario Nazionale 2005 (to A.P.). S.C. is supported by the Giovanni Armenise-Harvard Foundation and by the Italian Association for Cancer Research.
D.O., D.A., and F.R. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat Immunol. 2002;3:323–328. doi: 10.1038/ni0402-323. [DOI] [PubMed] [Google Scholar]
- Busslinger M, Nutt SL, Rolink AG. Lineage commitment in lymphopoiesis. Curr Opin Immunol. 2000;12:151–158. doi: 10.1016/s0952-7915(99)00065-5. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis: a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- van Oostveen J, Bijl J, Raaphorst F, Walboomers J, Meijer C. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia. 1999;13:1675–1690. doi: 10.1038/sj.leu.2401562. [DOI] [PubMed] [Google Scholar]
- Acampora D, Simeone A. The TINS Lecture: understanding the roles of Otx1 and Otx2 in the control of brain morphogenesis. Trends Neurosci. 1999;22:116–122. doi: 10.1016/s0166-2236(98)01387-3. [DOI] [PubMed] [Google Scholar]
- Puelles E, Acampora D, Lacroix E, Signore M, Annino A, Tuorto F, Filosa S, Corte G, Wurst W, Ang SL, Simeone A. Otx dose-dependent integrated control of antero-posterior and dorso-ventral patterning of midbrain. Nat Neurosci. 2003;6:453–460. doi: 10.1038/nn1037. [DOI] [PubMed] [Google Scholar]
- Simeone A, Puelles E, Acampora D. The Otx family. Curr Opin Genet Dev. 2002;12:409–415. doi: 10.1016/s0959-437x(02)00318-0. [DOI] [PubMed] [Google Scholar]
- Levantini E, Giorgetti A, Cerisoli F, Traggiai E, Guidi A, Martin R, Acampora D, Aplan PD, Keller G, Simeone A, Iscove NN, Hoang T, Magli MC. Unsuspected role of the brain morphogenetic gene Otx1 in hematopoiesis. Proc Natl Acad Sci USA. 2003;100:10299–10303. doi: 10.1073/pnas.1734071100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Eberhart CG, Riggins GJ. Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 2005;65:703–707. [PubMed] [Google Scholar]
- Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, Lengauer C, Cummins JM, Velculescu VE, Fults DW, McLendon RE, Bigner DD, Yan H. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–924. [PubMed] [Google Scholar]
- de Haas T, Oussoren E, Grajkowska W, Perek-Polnik M, Popovic M, Zadravec-Zaletel L, Perera M, Corte G, Wirths O, van Sluis P, Pietsch T, Troost D, Baas F, Versteeg R, Kool M. OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol. 2006;65:176–186. doi: 10.1097/01.jnen.0000199576.70923.8a. [DOI] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- Gattei V, Degan M, Gloghini A, De Iuliis A, Improta S, Rossi FM, Aldinucci D, Perin V, Serraino D, Babare R, Zagonel V, Gruss HJ, Carbone A, Pinto A. CD30 ligand is frequently expressed in human hematopoietic malignancies of myeloid and lymphoid origin. Blood. 1997;89:2048–2059. [PubMed] [Google Scholar]
- Swerdlow SH CE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. Lyon: IARS Press,; World Health Organization Classification of Tumours of Hematopoietic and Lymphoid Tissues. 2008 [Google Scholar]
- Colomo L, Lopez-Guillermo A, Perales M, Rives S, Martinez A, Bosch F, Colomer D, Falini B, Montserrat E, Campo E. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood. 2003;101:78–84. doi: 10.1182/blood-2002-04-1286. [DOI] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Shi SR, Key ME, Kalra KL. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- Acampora D, Avantaggiato V, Tuorto F, Briata P, Corte G, Simeone A. Visceral endoderm-restricted translation of Otx1 mediates recovery of Otx2 requirements for specification of anterior neural plate and normal gastrulation. Development. 1998;125:5091–5104. doi: 10.1242/dev.125.24.5091. [DOI] [PubMed] [Google Scholar]
- Kastury K, Druck T, Huebner K, Barletta C, Acampora D, Simeone A, Faiella A, Boncinelli E. Chromosome locations of human EMX and OTX genes. Genomics. 1994;22:41–45. doi: 10.1006/geno.1994.1343. [DOI] [PubMed] [Google Scholar]
- Bea S, Colomo L, Lopez-Guillermo A, Salaverria I, Puig X, Pinyol M, Rives S, Montserrat E, Campo E. Clinicopathologic significance and prognostic value of chromosomal imbalances in diffuse large B-cell lymphomas. J Clin Oncol. 2004;22:3498–3506. doi: 10.1200/JCO.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Tagawa H, Kameoka Y, Kasugai Y, Karnan S, Kameoka J, Sasaki T, Morishima Y, Nakamura S, Seto M. Characterization of target genes at the 2p15–16 amplicon in diffuse large B-cell lymphoma. Cancer Sci. 2006;97:499–504. doi: 10.1111/j.1349-7006.2006.00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houldsworth J, Olshen AB, Cattoretti G, Donnelly GB, Teruya-Feldstein J, Qin J, Palanisamy N, Shen Y, Dyomina K, Petlakh M, Pan Q, Zelenetz AD, Dalla-Favera R, Chaganti RS. Relationship between REL amplification. REL function, and clinical and biologic features in diffuse large B-cell lymphomas. Blood. 2004;103:1862–1868. doi: 10.1182/blood-2003-04-1359. [DOI] [PubMed] [Google Scholar]
- Lossos IS. Molecular pathogenesis of diffuse large B-cell lymphoma. J Clin Oncol. 2005;23:6351–6357. doi: 10.1200/JCO.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Satterwhite E, Sonoki T, Willis TG, Harder L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, Schlegelberger B, Oscier DG, Siebert R, Tucker PW, Dyer MJ. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.