Abstract
Direct reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) can be achieved by overexpression of Oct4, Sox2, Klf4 and c-Myc transcription factors, but only a minority of donor somatic cells can be reprogrammed to pluripotency. Here we demonstrate that reprogramming is a continuous stochastic process where almost all donor cells eventually give rise to iPSCs upon continued growth and transcription factor expression. Additional inhibition the p53/p21 pathway or overexpression of Lin28 increased the cell division rate and resulted in an accelerated kinetics of iPSC formation that was directly proportional to the increase in cell proliferation. In contrast, Nanog overexpression accelerated reprogramming in a predominantly cell division rate independent manner. Quantitative analyses define distinct cell division rate dependent and independent modes for accelerating the stochastic course of reprogramming, and suggest that the number of cell divisions is a key parameter driving epigenetic reprogramming to pluripotency.
Quantifying the efficiency and timescales of crucial events occurring during in vitro reprogramming to pluripotency1–5 has been problematic due to the cellular and genetic heterogeneity of de novo infected somatic cells6,7. To circumvent the need for virus-mediated transduction and reduce the heterogeneity of reprogramming factor expression, a “secondary” reprogramming transgenic system was devised where all somatic cells carry the same integration pattern of drug inducible Oct4, Sox2, Klf4 and c-Myc (OSKM) viral transgenes8–11. Although reprogramming of somatic cells from secondary mice was two orders of magnitude higher than in freshly infected somatic cells, only 1–20% of the induced cells generated iPSCs after 3–4 weeks of factor expression9,10,12,13. Further, partially reprogrammed “intermediate” cell lines have been derived in different experimental settings, some of which can give rise much later to fully reprogrammed iPSCs either spontaneously or upon additional manipulations14,15. Given that the timescale of several weeks and relatively low efficiencies persist even after controlling for adequate OSKM transgene expression, these studies left important questions relevant to the basic mechanisms of epigenetic reprogramming unresolved: how does the reprogramming process progress over time and what happens to the majority of the cells that do not become reprogrammed upon continued cell growth and expression of the reprogramming factors? Why do some somatic cells, that circumvent senescence or apoptosis induced by OSKM, convert into iPSCs earlier than others? Do all adult donor cells expressing OSKM reprogramming factors eventually give rise to iPSCs or would this be achieved only upon additional genetic or small molecule manipulation? Is high reprogramming efficiency restricted to non-fully differentiated or adult stem cells6,7,16,17?
Models to account for the reprogramming process fall into two categories (Fig. 1). “Deterministic” models posit that either “all” (i) or only a subset of “elite” or “stem-like” cells (ii) within a donor population have the potential to generate iPSCs and are reprogrammed with a fixed latency. We define “latency” as the absolute time or the number of cell divisions that an individual donor cell undergoes until it gives rise to a daughter iPSC. “Stochastic” models posit that most if not all (iii) or only a subset of “elite” somatic cells (iv) within a donor population have the potential to generate iPSCs albeit with different latencies. To gain insight into the mechanism of reprogramming and to address some of these questions, we have characterized the reprogramming efficiency and kinetics of over 1000 somatic cell derived monoclonal populations expressing OSKM over an extended period of time and quantitatively defined distinct modes for changing the course of the reprogramming process upon additional genetic perturbations.
Figure 1. Models of progressing to a pluripotent state during direct reprogramming.
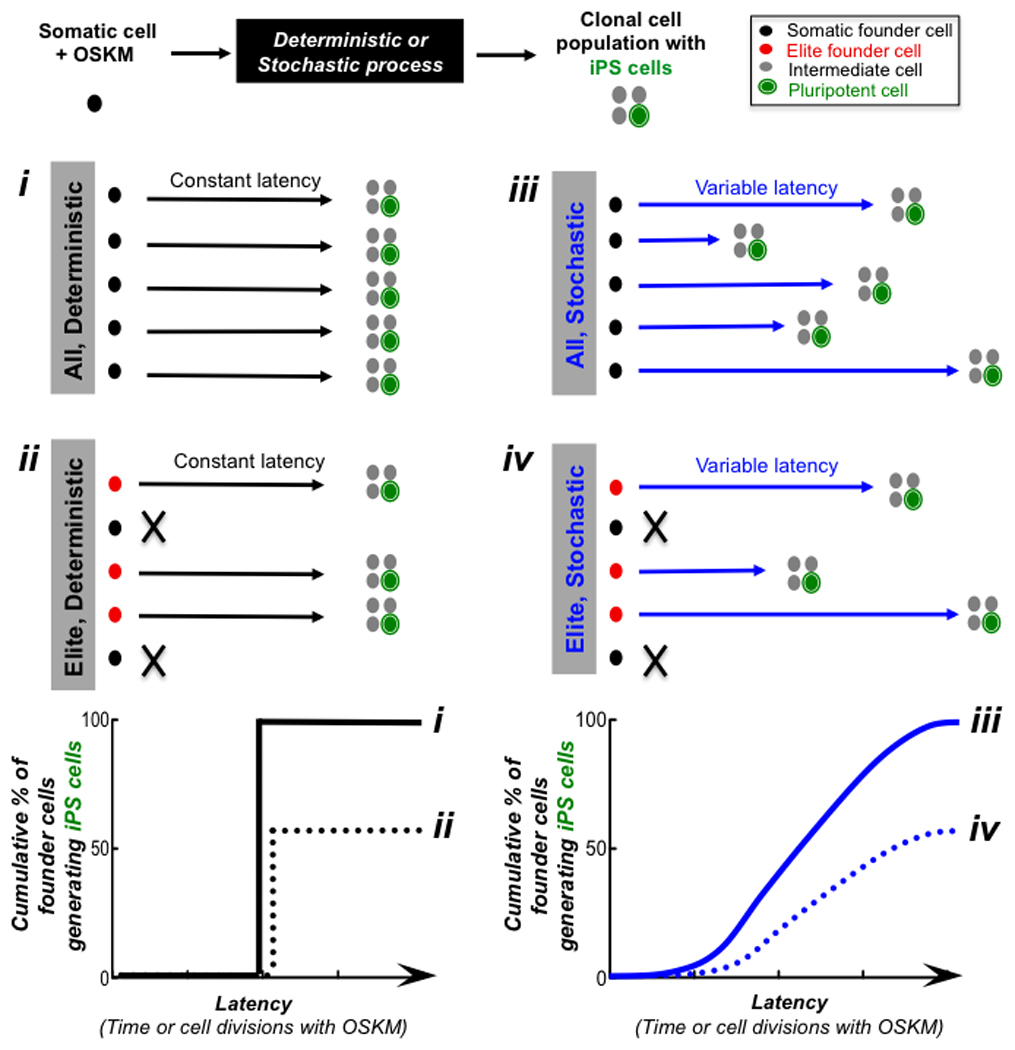
Four different models (i–iv) to account for the latency of donor somatic cells in progressing towards the induced pluripotent stem cell (iPSC) state following the expression of OSKM reprogramming factors. Latency can be measured in units of absolute time or cell divisions until the first iPSC is generated from a monoclonal population. Graphs display the general shape of the reprogramming kinetics in the different scenarios. Note that elite models do not necessarily reprogram more slowly as shown in the bottom plots.
Reprogramming of monoclonal populations
We followed the reprogramming of individual somatic donor cells and studied their potential to generate iPSCs. Unlike fibroblasts, B cell lineage-committed cells at the early Pre-B cell stage can be efficiently cloned as single cells immediately after isolation and were used as a defined, homogenous starting cell population for reprogramming into iPSCs. The NGFP1 iPSC line was generated by infecting fibroblasts from Nanog-GFP reporter mice with Doxycycline (DOX)-inducible lentiviral vectors encoding the OSKM transcription factors and injected into host blastocysts to generate “secondary” chimeras (Fig. 2a)9,18,19. NGFP1 derived secondary Pre-B cells were single cell sorted into individual wells and exhibited high cloning efficiency on DOX (>80%). Populations were serially passaged and monitored weekly for reactivation of the endogenous Nanog-GFP knock-in reporter (Supplementary Fig. 1), which represents one of the final events during reprogramming4,20–22. We defined “reprogramming efficiency” as the long-term potential of a cell to generate iPS daughter cells. A detection value of >0.5% for Nanog-GFP+ cells per well reproducibly allowed for stable derivation of Nanog-GFP+ iPSCs upon DOX withdrawal (Supplementary Fig. 2), and was set as the minimal threshold for defining positive detection of iPSCs in clonal populations.
Figure 2. Long-term analysis of reprogramming monoclonal populations.
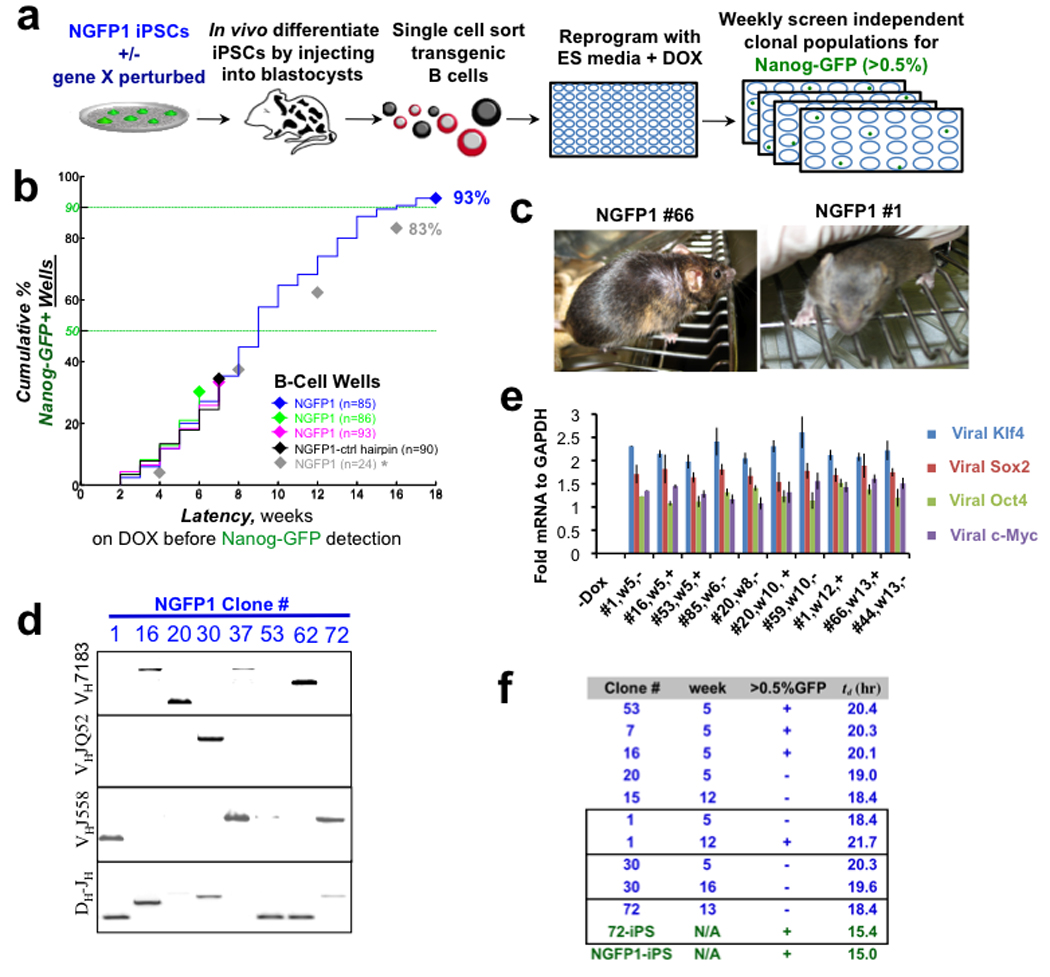
a, Schematic of experiments. b, Reprogramming of Pre-B cell monoclonal populations measured as the cumulative number of wells that became Nanog-GFP+. n indicates number of populations monitored. Asterisk indicates flow cytometry for GFP detection was performed every 4 weeks. c, Chimeric mice with agouti coat color from iPSCs derived after 12–13 weeks of DOX. d, Heavy chain rearrangements in iPSCs. e, Relative transgene induction levels of monoclonal populations on DOX. Error bars indicate standard deviation (n=3). f, The population averaged doubling time, td, for each clonal population. Boxes delineate cases where the same clonal population was measured at different times during DOX induction. Lower two lines (green) represent subcloned iPSC lines. Clone labeling: clone #, weeks on DOX (w#), Nanog-GFP >0.5% status (+/−).
Nanog-GFP+ cells were detected after two weeks of DOX induction, with ~3–5% of the wells generating Nanog-GFP+ cells at 2 weeks10–12 (Fig. 2b). The remaining wells contained viable cells, could be propagated in the presence of DOX, and uniformly silenced hematopoietic surface markers (Supplementary Fig. 3)15,22. Upon extended culture, the fraction of wells with iPSCs gradually increased and by 18 weeks after DOX addition >92% of the wells had produced Nanog-GFP+ cells (Fig. 2b), demonstrating that most if not all donor cells have the potential to generate iPSCs. The reprogramming kinetics were reproducible in independent experiments (Fig. 2b) and were not restricted to B cells, as CD11b+ monocytes that showed comparable transgene induction levels generated iPSCs with similar kinetics and efficiency (Supplementary Fig. 4). Nanog-GFP+ cell populations selected at random gave rise to stable DOX-independent iPSC lines. Furthermore, all tested iPSC lines had normal karyotypes and generated teratomas and chimeras irrespective of whether they were derived at early or late time points during the process (Fig. 2c and Supplementary Fig. 5). The iPSC lines carried distinct genetic heavy chain rearrangements verifying their independent clonal origin (Fig. 2d). We assessed whether transgene expression levels or increased proliferation rate could underlie the well-to-well differences seen in the latency of reprogramming. The population-averaged cell doubling times (td) and transgene induction levels during the reprogramming process were similar in NGFP1 clonal populations irrespective of time on DOX or whether these populations contained a Nanog-GFP+ fraction (Fig. 2e–f).
These results suggest: 1) Reprogramming of somatic cells is a continuous stochastic process where nearly all somatic donor cells have the ability to give rise to iPSCs upon continuous passaging and expression of OSKM. 2) Although reprogrammed cells do not appear prior to 8–10 days of OSKM expression21,22, the time of DOX exposure or number of cell divisions achieved before a given clonal population generates iPSCs varies widely. 3) Our data are not consistent with an “elite component” in reprogramming, as most if not all lineage-committed B cells or monocytes are able to generate iPSCs rather than only a small fraction of putative somatic stem cells present in the donor cell population 7,12–13. 4) Finally, somatic cells reprogram with different latencies that cannot be predicted based on time of DOX exposure or proliferation rate, consistent with undefined stochastic events driving the process (model iii in Fig. 1).
Cell division rate and reprogramming
We next characterized parameters of the reprogramming process by introducing defined genetic perturbations. Recently, p53 inhibition has been shown to enhance the efficiency of iPSC formation from fibroblasts by direct viral infection or transient transfections protocols by reducing apoptosis following initial transgene induction16,23–27. We tested whether and how p53 inhibition would influence the reprogramming of secondary transgenic NGFP1 iPSC derived Pre-B cells that have a high single-cell cloning efficiency and stably grow in the presence of DOX without requiring additional immortalization and with only background apoptosis levels (Supplementary Fig. 6). NGFP1 iPSCs were infected with a constitutively expressed lentiviral vector encoding a siRNA hairpin for p53 (Supplementary Fig. 7)28. Infected cells were injected into host blastocysts and NGFP1-p53 knock-down (p53KD) B cells were single cell sorted and cultured in DOX. p53 inhibition did not alter transgene expression levels or affect the already residual levels of apoptosis (Fig. 3a, Supplementary Fig. 6,8), but rather shortened the cell population-averaged doubling time of p53KD cells in the presence of DOX by ~2-fold as compared to control NGFP1 derived cells (Fig. 3b, Supplementary Fig. 9). The kinetics of iPSC formation proceeded with a significantly accelerated rate, with 93% of the wells producing Nanog-GFP+ cells within 8 weeks of DOX, as compared to 17 weeks of the control cells (Fig. 3c). The iPSC lines were DOX independent, expressed pluripotency markers and generated teratomas and mouse chimeras (Supplementary Fig. 10).
Figure 3. Cell division rate dependent and independent acceleration of reprogramming.
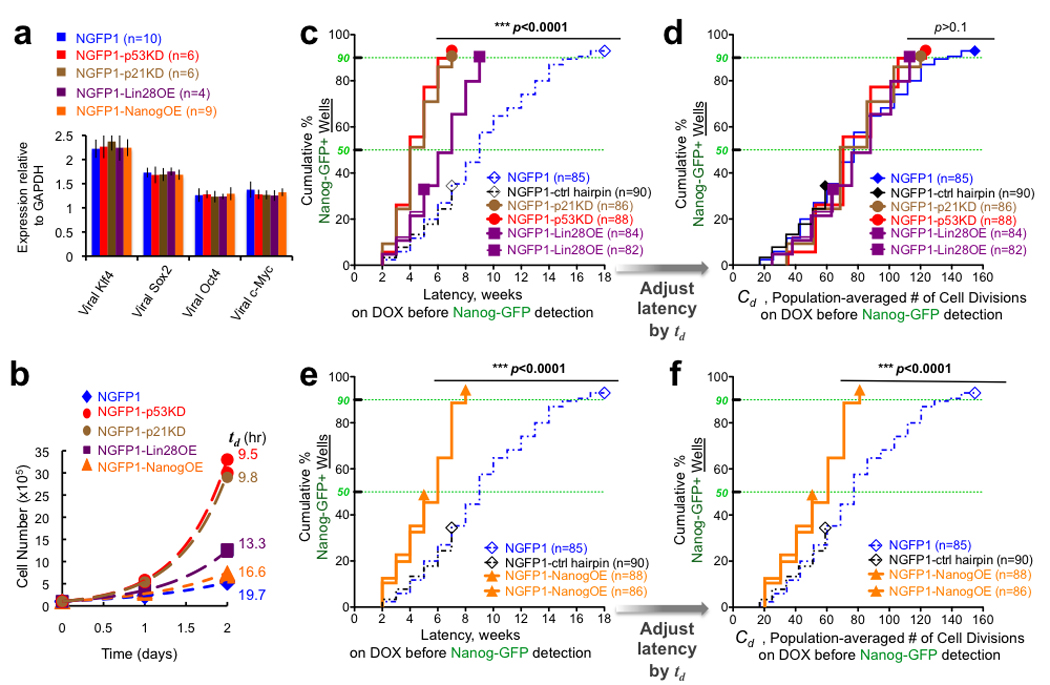
a, Average induction levels for transgenes in different NGFP1 cell populations. n indicates number of populations sampled per group, presented as mean ± s.d. b, Growth curves for cells on DOX. Exponential growth (dashed line) described the data well (R2=0.97–1.0), and the population-averaged doubling times (td) were calculated from these fits (Supplementary Fig. 9). c, As in Fig. 2b, latencies for reprogramming various clonal B cell derived populations. NGFP1-p53KD, NGFP1-p21KD, and NGFP1-Lin28OE wells were statistically distinct from the NGFP1 and NGFP1-control hairpin wells (p<0.0001, logrank test for dissimilarity). d, Rescaling time by td provides an estimate for the number of cell divisions occurring during latency. No statistical difference between groups was observed after rescaling time by td (p>0.1). e–f, As in c–d, but for NGFP1-NanogOE wells. n indicates number of populations monitored.
To assess whether the enhanced reprogramming could be attributed to p53 inhibition effect on proliferation rate, we estimated, based on the population-averaged doubling times measured throughout the process, how many cell divisions have occurred for each NGFP1 and NGFP1-p53KD clonal population during the latency period. Upon rescaling of latency by the doubling time (Fig. 3c,d), the cumulative fraction of wells generating iPSCs collapsed to the same statistically significant distribution with cell division number for both the NGFP1 and NGFP1-p53KD wells (log-rank test for dissimilarity, p=0.518). A similar analysis on NGFP1 and NGFP1-p53KD CD11b+ cell-derived clonal populations showed a comparable distribution (log-rank test for dissimilarity, p=0.209; Supplementary Fig. 11). Latencies were not normally distributed about the mean latency, but rather were better fit with a gamma distribution (Supplementary Fig. 12–13). Knockdown of p21 gene, a downstream effector of p53 which regulates cell cycle progression29, recapitulated the change in cell division rate and acceleration of reprogramming dynamics upon OSKM expression (Fig. 3a–d, Supplementary Fig. 6–9,14), further substantiating that change in cell division rate directly rescales the kinetics of reprogramming to distributions similar to OSKM alone. Finally, we studied the effect of lin28 overexpression on the reprogramming dynamics, as it enhances the reprogramming of human fibroblasts and acts as an oncogene by modulating the expression of cell cycle regulators5,30. NGFP1-Lin28OE (Lin28 overexpresser) derived B cell populations demonstrated an accelerated reprogramming kinetics that directly correlated with the observed ~30% increase in cell division rate on DOX (p=0.327, Fig. 3a–d). In summary, our data demonstrate that both p53/p21 inhibition or Lin28 overexpression accelerate the reprogramming process rather than enhancing overall efficiency24 as the cells divide more rapidly resulting in an increased cumulative probability for the stochastic events to occur earlier in time (Fig. 3a–d).
Cell division rate independent acceleration
We investigated whether reprogramming could be accelerated by mechanisms that are independent of cell proliferation rate. Nanog is a pluripotency factor expressed in the inner cell mass, and deriving ESCs and iPSCs requires the presence of functional endogenous nanog alleles31. Moreover, Nanog enhances the transfer of pluripotency by cell fusion and facilitates direct reprogramming of human cells5,32–35. Therefore, we aimed to investigate the effect of additional ectopic Nanog expression on the reprogramming kinetics by OSKM. B cells were derived from an NGFP1-NOE (Nanog overexpresser) iPSC line carrying a DOX inducible Nanog-encoding transgene (Supplementary Fig. 7). The presence of the transgene did not alter the expression of OKSM (Fig. 3a). Overall efficiency of iPSC formation was similar to that of the control cells but reprogramming occurred with a significantly accelerated kinetics with 94% of the wells producing pluripotent Nanog-GFP+ cells within 8 weeks of DOX (Fig. 3e, Supplementary Fig. 15–16). Nanog overexpression slightly increased the cell population-averaged doubling time of DOX-induced NGFP1-NOE cells compared to control NGFP1 derived reprogramming cells (Fig. 3b)36,37. However even after rescaling time on DOX by the change in doubling time (Fig. 3f), the cumulative fraction of wells generating iPSCs did not collapse to the same distribution with cell number (log-rank test for dissimilarity, p<0.0001). The median number of cell divisions before wells produced iPSCs was significantly reduced from about 70 cell divisions in the NGFP1, NGFP1-p21KD, and NGFP1-p53KD lines to 50 cell divisions in the NGFP1-NOE wells (Fig. 3d,f, Supplementary Fig. 12d). These results suggest that Nanog over expression accelerates the reprogramming kinetics by cell intrinsic mechanisms that are independent of an altered cell proliferation rate.
Numerical modeling of reprogramming
We utilized our datasets to conduct simulations of the stochastic reprogramming process occurring in individual cells. To assess the intrinsic rate of reprogramming per cell, we tested whether a simple stochastic model would describe our observations. The model considers the reprogramming of B cells to occur as a one-step process with a constant cell-intrinsic rate k (Fig. 4a). Because iPSCs were adherent and selectively retained throughout the culturing procedures, the latency measured for each well was representative of the timing at which the first B-cell in the population reprogrammed plus a time delay, tp, during which the daughter cells needed to grow to reach the detection threshold. At any given time t, the number of cells in each well, N(t), scales the rate at which the first reprogramming event takes place, and the cumulative probability distribution of reprogramming times is P(t+tp)≅1–exp(−kτ), where τ is the “population-rescaled time,” . Interpreting the experimental results in terms of this rescaled version of time allowed to separate the contributions of population size and cell intrinsic reprogramming rate (Fig. 4). Cell division rate can control the population size, N, and thus rescales time, affecting the observed rate of reprogramming. For example, if the cell division rate and culturing procedures were modified to an effective population size of 1000 cells in each well rather than ~106 cells per well (Fig. 4b–c), longer times would have been necessary to reach >90% reprogrammed wells.
Figure 4. Quantitative analysis of cell reprogramming.
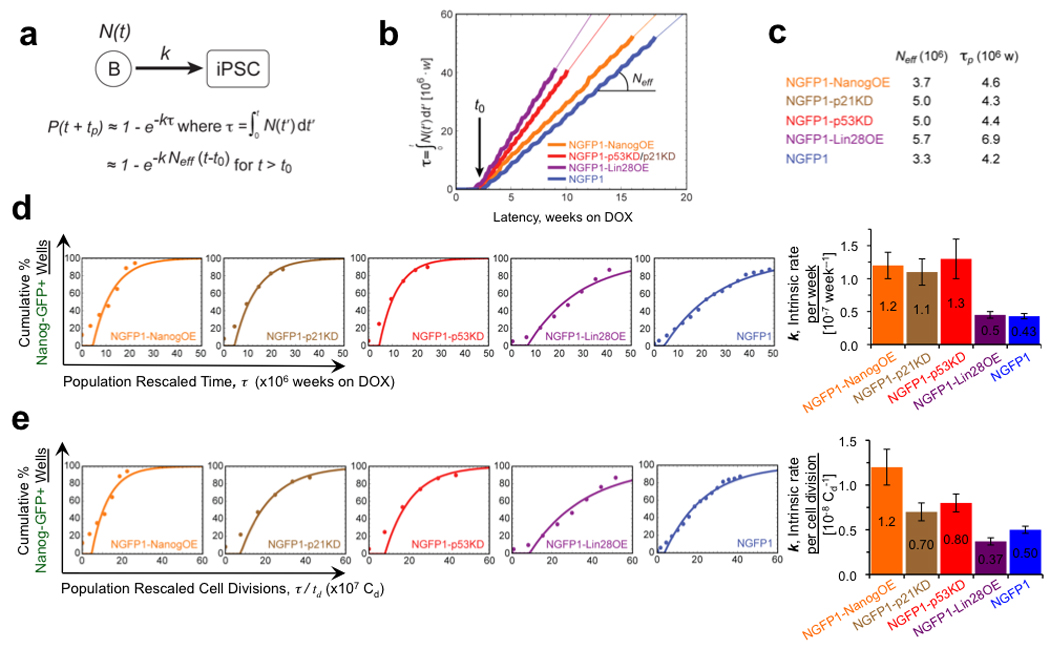
a, Stochastic model summary. Sequential replating of individual wells during each experiment establishes that, after a time t0 representative of the time at which the replatings started, each experiment can be described in terms of a population of an effective size, Neff. b, Estimate of the population rescaled time, τ, throughout each experiment. After t0, population dynamics are effectively described by a fixed population of size, Neff. NGFP1-p53KD and NGFP1-p21KD have similar dynamics. c, Neff and the population-rescaled average proliferation times, τp, estimated as the population-rescaled time necessary for one iPSC to reach the detection threshold [τp=td,iNefflog2(ρNeff), where ρ is the detection threshold and td,i is the doubling time of iPSCs]. d, Cumulative percentage of Nanog-GFP+ wells as a function of τ, and best fits according to the proposed model. Modeling results are in thick lines, and experiments are in dots. Right graph indicates best fit estimates of the cell-intrinsic rate k expressed in terms of weeks. e, As in d, but per population-rescaled cell divisions, τ/td, instead of per τ units. td is the doubling time of the populations. Error bars indicate 95% confidence intervals.
Using cell population dynamics to account for the growth and culturing procedures used across the different NGFP1 lines (Fig. 4b–c), we calculated the population rescaled time and fit the cell-intrinsic rate k using a likelihood maximization approach (Fig. 4d–e, Supplementary Fig. 17). Reasonable agreement was seen between best fits and the experiments. Further, to take into account fluctuations in cell division times, number of cells and potential loss of iPSCs during culturing in addition to the stochasticity in the cell-intrinsic reprogramming process, we implemented a detailed computer simulation of each experiment (Supplementary Fig. 18). Simulation results were consistent with those achieved using the analytical approach (Fig. 4d–e). The inferred intrinsic reprogramming rates per cell division for NGFP1-p53KD and NGFP1-p21KD cells were similar to NGFP1 cells (Fig. 4e) suggesting that p53/p21 pathway inhibition in our system accelerates reprogramming through a predominately cell-division rate dependent mechanism. The slightly higher cell-intrinsic rates in some of our modeling results for p53/p21 inhibition (Fig. 4d–e, Supplementary Fig. 17–18) may reflect a modest acceleration due to cell-division rate independent mechanisms and we cannot exclude that inhibition of p53/p21 pathway slightly enhances reprogramming by additional mechanisms as well as in different experimental systems. The acceleration observed for NGFP1-Lin28OE cells occurred predominantly due to a larger effective population size resulting from a faster cell division rate on DOX when following a culturing procedure similar to that used for NGFP1 cells (Fig. 4b–c). Only in the case of NGFP1-NanogOE cells, higher than 1.75–2 fold intrinsic reprogramming rates per cell division compared to NGFP1 cells was robustly obtained in all modeling approaches (Fig. 4e, Supplementary Fig. 17–18), supporting the hypothesis that Nanog overexpression accelerates reprogramming through a predominately cell-division rate independent mechanism. Future characterization of how Nanog molecularly orchestrates the reestablishment of the core pluripotency circuitry is of great interest.
Discussion
We utilized a highly defined quantitative system for analyzing the reprogramming process and suggest that yet to be defined rate-limiting stochastic events occur as a function of cell division before the fully reprogrammed pluripotent state is attained. Upon OSKM induction in our system, most if not all monocytes and lineage committed B cells harboring genetic rearrangements have the potential to generate iPSCs albeit with very different latencies (model iii, Fig. 1). These results are inconsistent with models in which iPSCs preferentially arise from a particular epigenetic state in the donor cell population, such as a progenitor or an adult stem cell (models ii and iv, Fig. 1). The dynamics of direct in vitro reprogramming to pluripotency by OSKM are consistent with a continuous stochastic process, in which the conversion from a somatic cell to an iPSC can be thought of as a drift in cell state. This cell state can be defined by a gene expression or epigenetic pattern, whose fluctuations drive the conversion to an iPSC. These fluctuations are likely due to cell-to-cell extrinsic heterogeneity or the inherent stochastic nature of gene expression or regulatory signaling processes38–40. In the simplest scenario, the process could be modeled as a one-step rate limiting transition characterized by a cell-intrinsic rate, and we found that this model describes reasonably the observed dynamics across all of the different cell lines. Our model relies on inferring a single cell-intrinsic rate averaged over the entire course of reprogramming, though the model does not optimally describe the reprogramming behavior at early times following transgene induction (Fig. 4). This deviation between modeling and experimental results may indicate that the cell-intrinsic rate changes over the course of reprogramming within clonal populations. Intra-clonal monitoring of reprogramming dynamics at various points throughout the process could be used to add further mechanistic detail to the model, and to decipher the identity and sequence of rate limiting step(s) and epigenetic changes that occur during reprogramming.
Quantitative analysis of the differences in reprogramming latency upon different genetic perturbations defined two distinct modes of accelerating the reprogramming process (Fig. 5, Supplementary Fig. 19). In the “cell division rate dependent” mode, the cumulative probability for successful reprogramming is higher and can be achieved earlier in time and directly proportional to the augmentation in cell division rate, whereas in the “cell division rate independent” mode, reprogramming acceleration occurs over a lower average number of cell divisions. Notably, the different modes need not to be mutually exclusive as certain perturbations could enhance or inhibit reprogramming via both cell proliferation dependent and independent effects. Recent studies failed to attain correlation between cell division rate and reprogramming efficiency, however these studies relied on measuring proliferation in the absence of OSKM transgenes16 or by inducing proliferation with hematopoietic cytokines for which the receptors are rapidly silenced following transgene induction13. Our experimental and modeling data suggest that close monitoring of transgene induction, plating efficiency, cell proliferation, and changes in population size throughout the experiments are needed to gain insight into the stochastic process of reprogramming. We consider two possibilities for the ability of increased cell division rate to accelerate and drive the kinetics of the reprogramming process: 1) cell division could amplify the number of daughter cells from partially reprogrammed cells where each resulting individual cell has an independent probability of progressing towards becoming an iPSC, and 2) nuclear changes during cell division may facilitate the acquisition of epigenetic marks such as DNA and histone modifications that allow the re-establishment of the core transcriptional circuitry that stabilizes pluripotency6,9,41,42.
Figure 5. Distinct modes for accelerating reprogramming to pluripotency.
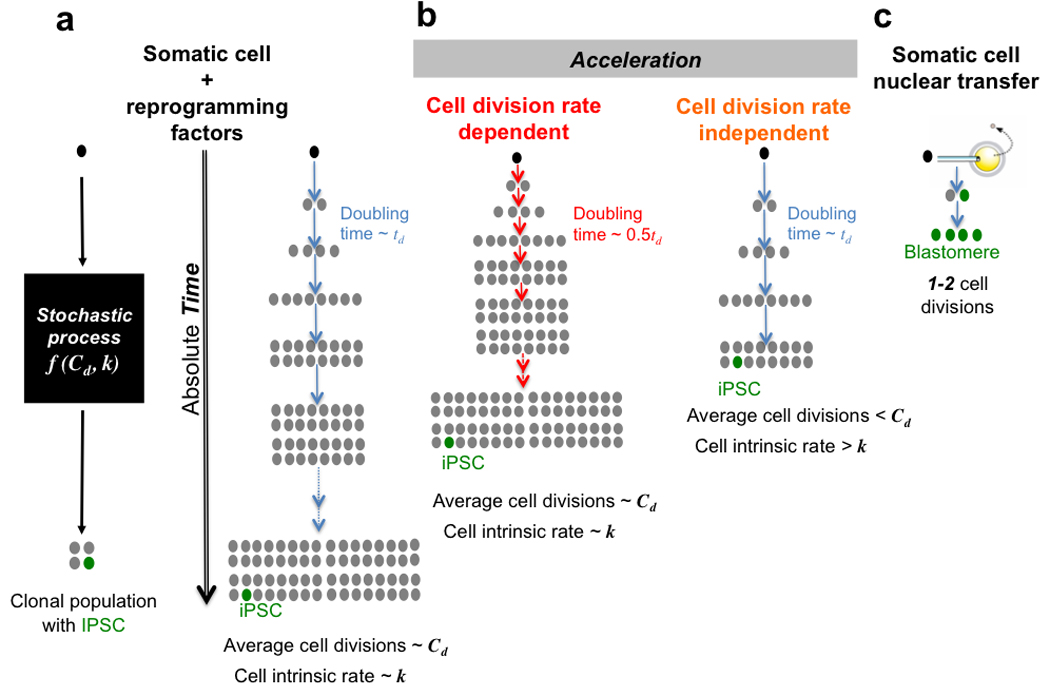
a, Nearly all donor cells can give rise to iPSCs via a stochastic process. Two parameters characterize the kinetics of the process: the average number of cell divisions required, Cd, and the cell-intrinsic reprogramming rate per cell division, k. b, Accelerating reprogramming in a cell division rate-dependent manner still requires as many divisions as the unaccelerated reference scenario (i.e., still Cd on average) but occurs earlier in time because cells divide faster, whereas in the cell division rate-independent mode, the cell-intrinsic rate reflecting the occurrence of unknown stochastic event(s) is enhanced and reprogramming is achieved within a lower average number of divisions (<<Cd). c, In comparison, somatic cell nuclear transfer can reprogram within 1–2 cell divisions.
iPSCs can be derived by various combinations of transcription factors and/or small molecules3,5,43,44. Our study identifies the average number of cell divisions required to give rise to an iPSCs at a particular efficiency as a key parameter that may be affected by different reprogramming strategies. After nuclear transfer, the pluripotency gene Oct4 of the somatic nucleus is reactivated in the cloned embryo within 1–2 cell divisions (Fig. 5)42,45,46 suggesting that the egg cytoplasm carries as-of-yet undefined determinants that accomplish robust reprogramming within very few cell divisions. These observations raise questions of whether direct reprogramming recipes devised so far are “reductionist” in nature, and whether the conversion into iPSCs can be enhanced to enable complete in vitro reprogramming within only a few cell divisions (e.g. optimizing stoichiometry, small molecules, oocyte derived transcript libraries).
Methods summary
The NGFP1 iPSC line was derived after culturing the infected MEFs carrying ROSA26-M2rtTA mice and Nanog-GFP alleles with Doxycycline (DOX)-inducible lentiviruses encoding Oct4, Klf4, c-Myc and Sox2 cDNA driven by the TetO/CMV promoter9. To generate the different genetically perturbed NGFP1 subclones, 50,000 NGFP1 cells were infected with pSicoR vector28 encoding a hairpin for the p53 or p21 tumor suppressor genes or with DOX inducible lentiviruses encoding Nanog or Lin28 cDNAs. To generate mouse chimeras iPSCs were injected into diploid blastocysts that were initially harvested 94–98 hours after hormone injection and placed in a drop of DMEM with 15% FCS under mineral oil. Pre-BCR+ (Igµ+ IgK− IgL−) early-Pre B cells were single cell sorted from 3–5 week old chimeras and were plated in gelatinized and irradiated OP9 coated wells with ES media supplemented with DOX (4 µg/ml). IL-7 and SCF (10ng/ml each, Peprotech) were supplemented to the media for the first 1–2 weeks to optimize cloning efficiency in the presence of DOX. After 2 weeks of DOX induction, cultures were trypsinized every week and passaged on gelatinized plates in ES cell medium + DOX. Populations were defined positive for iPSC generation when >0.5% of the 10,000 adherent/semi-adherent screened cells by FACS were GFP+. NGFP1-p53KD and p21KD derived cells were passaged twice a week (versus once a week at the time of GFP assays for the other groups) to avoid over-confluence in the culture wells due to their accelerated proliferation rate. The reprogramming experiments were stopped at different time points and all non-reprogrammed populations were viable and demonstrated robust growth on DOX upon ending each experiment. For teratoma generation, 2×106 iPSCs were injected subcutaneously into both flanks of recipient SCID mice, and tumors were harvested for sectioning 3–6 weeks after initial injection.
Methods
Pluripotent lines and viral infections
Mouse embryonic fibroblasts (MEFs) used to derive primary induced pluripotent stem cell (iPSC) lines by infections with inducible lentiviruses were harvested at 13.5 days post coitum from F1 mating between ROSA26-M2rtTA and Nanog-GFP mice9. Lentiviral preparation and infection with Doxycycline (DOX)-inducible lentiviruses encoding Oct4, Klf4, c-Myc and Sox2 cDNAs driven by the TetO/CMV promoter, were previously described21. The NGFP1 iPSC line used in this study was derived after culturing the infected MEFs with DOX, and grew stably in culture independent of DOX. To generate NGFP1-p53KD and p21KD clonal cell lines, 50,000 NGFP1 cells were infected with the pSicoR-PGK-puro vector encoding a hairpin for mouse p21 (targeting sequence: GCAGATTGGTCTTCTGCAA) and a previously described specific hairpin for the mouse p53 tumor suppressor gene28. Hairpin against the CD8 sequence was used as control where indicated. 10 µg of lentiviral vector and packaging vectors were co-transfected in 293T cells by using the FuGENE 6 reagent (Roche Diagnostics). Supernatants were collected 36–48 hours after transfection, filtered through a 0.45-µm filter, and used to infect the iPSCs for 48 hours. Afterwards, the cells were trypsinized, plated at low densities and subcloned to test for knockdown specificity (NGFP1 already contains a puromycin selection cassette in the ROSA26 locus restricting our ability to use puromycin as a marker for infected cells). To verify integration of PsicoR lentiviral vectors the following oligos were used: Forward, CCCGGTTAATTTGCATATAATATTTC; and Reverse, CATGATACAAAGGCATTAAAGCAG. cDNAs encoding Nanog and Lin28 transcripts where cloned into EcoRI site of TetO-FUW lentiviral vectors and were used to infect NGFP1 iPSC. Integration for TetO-Nanog and TetO-Lin28 lentiviral vectors was verified by PCR and southern analysis on genomic DNA using the EcoRI digested insert as a probe (data not shown). A NGFP1 subclone that demonstrated >95% constitutive knockdown of p53 (Trp53) or p21 genes products, termed NGFP1-p53KD or NGFP1-p21KD, were used in the subsequent experiments. iPSCs were cultured in DME containing 15% fetal calf serum (FCS), leukemia inhibiting factor (LIF), penicillin/streptomycin, L-glutamine, beta-mercaptoethanol and nonessential amino acids. Chromosomal karyotyping of iPSC lines was performed by Cell Line Genetics on 20 G-banded metaphase cells from each line tested.
Chimera and teratoma formation
iPS cells were injected in C57B6 X 129Sv F1 Rag2−/− or BDF2 diploid blastocysts that were initially harvested 94–98 hours after hormone injection and placed in a drop of DMEM with 15% FCS under mineral oil. A flat-tip microinjection pipette with an internal diameter of 1.2–1.5 mm was used for iPSC injection (using a Piezo micromanipulator). A controlled number of cells was injected into the blastocyst cavity. After injection, blastocysts were returned to KSOM media (Invitrogen) and placed at 37°C until transferred to recipient females. Ten to fifteen injected blastocysts were transferred to each uterine horn of 2.5 days post coitum pseudo-pregnant females. To recover full-term pups, recipient mothers were sacrificed at 19.5 days post coitum. For teratoma generation, 2×106 iPSCs were injected subcutaneously into both flanks of recipient SCID mice, and tumors were harvested for sectioning 3–6 weeks after initial injection.
Reprogramming into iPSCs
Transgenic Pre-BCR+ (Igµ+ IgK− IgL−) early Pre-B cells, which are not generated in Rag2−/− mice due to their inability to undergo heavy and light chain rearrangements, were isolated from bone marrow of 3–5 week old Rag2−/− NGFP1 chimeras and single cell sorted into 96 well plates. This choice of host ensured that any isolated B cells were derived from the injected iPSCs, and not from the host blastocysts. In certain experiments, we labeled the NGFP1-iPS cell clone with a lentivirus constitutively expressing dTomato (a kind gift from K. Hochedlinger) and injected cells into BDF2 blastocysts to produce chimeric mice, and utilized the dTomato as a marker of transgenic cells. One of the experimental replicates was performed on B220+CD25+ Pro-B cells isolated from reprogrammable mice carry the same set of transgenes11. Cells were plated in gelatinized and irradiated (3000 Rad) OP9 coated wells in ES medium containing DOX (4 µg/ml). IL-7 and SCF (10 ng/ml each, Peprotech) were added to the medium for the first 1–2 weeks to optimize cloning efficiency in the presence of DOX (the plating efficiency of Pre-B cells to grow as single cells was > 80%). Cells on DOX did not exhibit contact inhibition and grew both on gelatin and in suspension. After 2 weeks of DOX induction, cultures were trypsinized every week and passaged on gelatinized plates in ES medium + DOX. Populations were defined as being positive for iPSC generation when >0.5% of the adherent/semi-adherent screened cells were GFP+. Upon detection of a GFP+ fraction, further flow cytometric (FACS) assaying of the clonal population was discontinued and, in all cases, stable DOX independent iPSC lines were derived by growing the cells in the absence of DOX (which allows the iPSCs to overgrow transgene-dependent, partially reprogrammed GFP- cell in the population). A fraction of ~250,000 cells, including a quarter of all fully-adherent cells, was replated at the end of each week (beginning at week 2) for further follow-up analysis. By including adherent cells in the culture plate upon passaging, we consistently retained all iPSCs generated during a given culture period, as no iPSCs were detected by FACS in the non-adherent fraction (Supplementary Fig. 1). NGFP1-p53KD and NGFP1-p21KD derived cells were passaged and replated twice a week (versus once a week at the time of GFP assays for other cells) to avoid over-confluence in the culture wells due to their higher proliferation rate. Population-averaged doubling time for reprogramming and iPSC populations was determined by plating 105 cells, and counting total cell number after 24 and 48 hours in duplicates. Numbers of cells as a function of time were fit using exponential growth regression in Excel (R2 ranging from 0.97 to 1.00). The CD11b+ myeloid fraction was sorted from spleen and cells were plated in gelatinized and irradiated OP9 coated 96 wells with ES medium supplemented with DOX (4 µg/ml). M-CSF, Flt3L, LPS and SCF (10 ng/ml, Peprotech) was added to the medium for the first 2 weeks to boost plating efficiency. Wild-type CD11b+ cells were obtained from “reprogrammable” transgenic mice carrying identical DOX inducible copies of the reprogramming factors Oct4, Sox2, Klf4 and c-Myc, Rosa26-M2rtTA and a Nanog-GFP knock-in reporter. Unlike iPSC-chimeras, all cells in these “reprogrammable” mice carrying the same set of transgenes11. However, unlike for Pre-B cells which are not found in Rag2−/− hosts and thus all isolated cells were of transgenic origin, CD11b+ cells were isolated from reprogrammable” mice carry the same set of transgenes11 or from dTomato labeled chimeric donor mice. V(D)J-IgH rearrangements were amplified from genomic DNA samples by PCR using degenerate primer sets as described9.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 20 minutes at 25 °C, washed 3 times with PBS and blocked for 15 min with 5% FBS in PBS containing 0.1% Triton-X. After incubation with primary antibodies against Oct4 (Santa Cruz), Nanog (polyclonal rabbit, Bethyl) and SSEA1 (monoclonal mouse, Developmental Studies Hybridoma Bank) for 1 h in 1% FBS in PBS containing 0.1% Triton-X, cells were washed 3 times with PBS and incubated with fluorophore-labeled appropriate secondary antibodies purchased from Jackson Immunoresearch. Specimens were analyzed on an Olympus Fluorescence microscope, and images were acquired with a Zeiss Axiocam camera.
Quantitative RT-PCR
Total RNA was isolated using Rneasy Kit (Qiagen). Three micrograms of total RNA was treated with DNase I to remove potential contamination of genomic DNA using a DNA Free RNA kit (Zymo Research, Orange, CA). One microgram of DNase I-treated RNA was reverse transcribed using a First Strand Synthesis kit (Invitrogen) and ultimately resuspended in 100 ul of water. Quantitative PCR analysis was performed in triplicate using 1/50 of the reverse transcription reaction in an ABI Prism 7000 (Applied Biosystems, Foster City, CA) with Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen). Error bars indicate standard deviation of triplicate measurements for each clone. Primers used for transgene encoded amplification were as follows: c-Myc: Forward, 5’-ACCTAACTCGAGGAGGAGCTGG-3’ and Reverse, 5’-TCCACATAGCGTAAAAGGAGC-3’; Klf4: Forward, 5’-ACACTGTCTTCCCACGAGGG-3’ and Reverse, 5’-GGCATTAAAGCAGCGTATCCA-3’; Sox2: Forward, 5’-CATTAACGGCACACTGCCC-3’ and Reverse, 5’-GGCATTAAAGCAGCGTATCCA-3’; Oct4: Forward, 5’-AGCCTGGCCTGTCTGTCACTC-3’ and Reverse, 5’-GGCATTAAAGCAGCGTATCCA-3’. Nanog: Forward 5’-ACATGCAACCTGAAGACGTG-3’ and Reverse; 5’-CACATAGCGTAAAAGGAGCAA-3’. To ensure equal loading of cDNA into RT reactions, GAPDH mRNA was amplified using the following primers: Forward, 5’-TTCACCACCATGGAGAAGGC-3’; and Reverse, 5’-CCCTTTTGGCTCCACCCT-3’. Data were extracted from the linear range of amplification.
Antibodies and apoptosis measurement assays
Fluorescently conjugated antibodies (PE, FITC, Cy-Chrome or APC labeled) were used for flow cytometric analysis, and for cell sorting: anti-CD11b+, Pre-BCR, IgK, IgL, CD19, B220, CD45.2 and IL7R antibodies (BD-Biosciences) were used. Enrichment for CD11bhigh cells by using CD11b magnetic bead isolation kit was use prior to sorting. Cell sorting was performed by using FACS-Aria (BD-Biosciences), and consistently achieved cell sorting purity of >99%. Antibodies for western blot analysis: anti-mouse p53 (BAF1355, R&D systems), anti-β actin (ab8226, Abcam) and anti-p21cip (clone C-19, Santa Cruz Biotechnology). To determine the rate of apoptosis, samples were stained with annexin V staining kit (BD Biosciences) and propidium iodide (PI) according to the manufacturer's recommendations and analyzed by flow cytometer. In addition, samples were subjected to the Tdt-mediated dUTP-biotin nick end labeling (TUNEL) test for apoptotic cells by using flow cytometry based MEBSTAIN apoptosis kit (MBL).
Non-parametric statistical analysis
The latency of iPSC appearance of each well was used to generate a survival curve for each experiment. Latency was considered either as a function of absolute time (i.e., the weeks on DOX until GFP was detected) or as a function of the population-averaged number of cell divisions during latency (i.e., latency in time divided by doubling time). Any wells that failed to generate Nanog-GFP+ cells at the end of each experiment were considered to be censored, and are shown with solid geometric shapes in Fig. 2–3. Histograms of the fraction of cells at initial GFP detection were performed using Prism 5 (version 5.0b; Graphpad Software, Inc.). Using the censored latency data sets, the logrank (Mantel-Cox) test, a nonparametric statistical test appropriate for right-censored data, was used to test the null-hypothesis that survival functions do not differ across groups. In contrast to parametric analyses, this test does not require knowledge about the shape of the survival curve or the distribution of survival times. Analysis was performed using Prism 5 (version 5.0b; Graphpad Software, Inc.), and two-tailed p-values. p-values above 0.05 indicate that the latencies between the two groups were similar (i.e., the treatment did not change survival; accept the null hypothesis at a 95% confidence level), while p-values less than 0.05 indicate that latencies between groups were not similar (i.e., survival functions differ; reject the null hypothesis at a 95% confidence level).
Parametric statistical analysis
The censored latency data sets were also fit to several univariate probability distributions using maximum likelihood estimation via the ‘dfitool’ in Matlab (The Mathworks, Inc.). For optimal univariate distribution fits, the chi-squared goodness-of-fit statistic, χ2, was used to assess the quality of each fit. Gamma distributions had the lowest χ2 of any fit distribution (see Supplementary Fig. 12).
Computational simulation
Simulations of the proposed model were generated using a hybrid scheme in which the size of each subpopulation (either B-cells or iPSC) was evolved by considering (a) stochastic Gillespie-like evolution of Poissonian growth dynamics for small population sizes, (b) deterministic evolution of population sizes using a time step of Δt = 0.001 weeks whenever the probability of generating a new cell in the time Δt exceeded 0.1, (c) Gillespie-like evolution of the reprogramming transition, (d) periodic replating, selection of iPSCs and observation of the fraction of reprogrammed cells according to the details of the experimental protocol followed in each experiment.
Supplementary Material
Acknowledgements
We would like to thank N. Maheshri, K. Wood, Y. Bugnanim, B. Carey, J. Cassady and F. Soldner for commenting on the manuscript; J. Dausmann, S. Markoulaki, P. Schorderet and M. Pawlak for conducting blastocysts injections and members of the Jaenisch lab for helpful discussions. R.J. is supported by grants from the NIH: RO1-HDO45022, R37-CA084198, RO1-CA087869. J.H. is supported by a postdoctoral fellowship from Helen Hay Whitney Foundation. K.S. is supported by the Society in Science: The Brano-Weiss fellowship. J.v.Z. is supported by the Human Frontiers Science Program. A.v.O. is supported by a National Institutes of Health (NIH) Director’s Pioneer award and an award from the National Cancer Institute (U54CA143874).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information contains 19 figures and is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. J.H., K.S. and R.J. conceived the ideas for this study and designed the experiments. J.H. conducted the reprogramming experiments and data collection. M.P.C. and C.J.L. assisted in construct preparation, transgene induction and blastocyst injections. K.S., B.P., J.v.Z. and A.v.O. performed statistical and numerical analyses, and also constructed the stochastic models. J.H., K.S. and R.J. wrote the manuscript with contributions from all other authors.
Author Information. Reprints and permissions information is available at www.nature.com/reprints.
Competing financial interests. R.J. is an advisor to Stemgent and a cofounder of Fate Therapeutics.
References
- 1.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318(5858):1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460(7251):49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 8.Hanna J, Carey BW, Jaenisch R. Reprogramming of Somatic Cell Identity. Cold Spring Harb Symp Quant Biol. 2008 doi: 10.1101/sqb.2008.73.025. [DOI] [PubMed] [Google Scholar]
- 9.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133(2):250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26(8):916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markoulaki S, et al. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat Biotechnol. 2009;27(2):169–171. doi: 10.1038/nbt.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna J, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4(6):513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eminli S, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;49:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sridharan R, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136(2):364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huangfu D, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev. 2008;22(22):3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc Natl Acad Sci U S A. 1993;90(10):4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 21.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2(2):151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2(3):230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23(18):2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1145–1148. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marion RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101(28):10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7(11):1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 30.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15(3):357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138(4):722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 33.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 34.Navarro P, et al. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321(5896):1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 35.Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441(7096):997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 36.Darr H, Mayshar Y, Benvenisty N. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 2006;133(6):1193–1201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- 37.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460(7251):118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 38.Huang S. Reprogramming cell fates: reconciling rarity with robustness. Bioessays. 2009;31(5):546–560. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- 39.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6(6):451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 40.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135(2):216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol. 2008;9(7):505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136(3):411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3(5):568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Boiani M, et al. Variable reprogramming of the pluripotent stem cell marker Oct4 in mouse clones: distinct developmental potentials in different culture environments. Stem Cells. 2005;23(8):1089–1104. doi: 10.1634/stemcells.2004-0352. [DOI] [PubMed] [Google Scholar]
- 46.Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447(7145):679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


