Abstract
The high molecular mass (HMM) PBPs are essential for bacterial cell wall biosynthesis, and are the lethal targets of the β–lactam antibiotics. When purified, the HMM PBPs give undetectable or weak enzyme activity. This has impeded efforts to develop assays for the HMM PBPs, and to develop new inhibitors for the HMM PBPs as HMM PBP targeted antibacterial agents. However, even when purified the HMM PBPs retain their ability to bind β-lactams. We describe here a fluorescently detected microtiter plate-based assay for inhibitor binding to the HMM PBPs based on competition with biotin-ampicillin (BIO-AMP) conjugate binding.
Keywords: β-lactam, assay, penicillin-binding protein, antibacterial, cell wall
INTRODUCTION
Penicillin-binding proteins (PBPs)2 are bacterial enzymes that catalyze the final steps in cell wall biosynthesis, and are the lethal targets of the β–lactam antibiotics (reviewed in [1-4]). Every bacterial species has multiple PBPs. For example, E. coli has eight classically known PBPs, labeled 1A, 1B, and 2-7. PBPs have molecular masses of 20-120 kDa and can be broadly divided into two groups, the high molecular mass (HMM) PBPs and the low molecular mass (LMM) PBPs [1]. HMM PBPs are essential for bacterial survival and are the lethal targets for β–lactam antibiotics, whereas LMM PBPs are non-essential for cell viability.
An enigmatic feature of the PBPs is that, while LMM PBPs give readily detectible activity against peptide substrates, purified HMM PBPs give either low or undetectable activity against natural or synthetic cell wall-related peptide substrates (reviewed in [5, 6]). This has impeded the development of convenient assays for the HMM PBPs. Two approaches which have had some success for demonstrating the activity of the HMM PBPs are the use of thiolester-based substrates [7, 8], and assays based on the use of lipid II [9-12], which is a precursor to the nascent peptidoglycan substrate of the PBPs. However, neither of these assays appear well suited for microtiter plate based high throughput assays – the thiolesters because of their high background rate of hydrolysis [7], and lipid II because of its difficult isolation [13] and synthesis [11, 12]. In an effort to circumvent the limitations of these and other HMM PBP assays, deSousa and coworkers have developed scintillation proximity assays to measure membrane associated peptidoglycan synthesis in E. coli membrane preparations [14-17], but these assays also appear difficult and cumbersome.
We describe here a general assay for screening and characterizing HMM PBP inhibitors. This approach is based on the fact that the HMM PBPs, essentially by definition, bind β–lactams. This assay uses a β–lactam-biotin conjugate (BIO-AMP) previously described for the detection of PBPs in Western Blots [18-20]. In the present study purified PBPs were immobilized onto microtiter plate wells, and labeled with BIO-AMP. Treatment of the BIO-AMP labeled PBP with a strepavidin-horse radish peroxidase (HRP) conjugate followed by a fluorogenic HRP substrate (Amplex Red) allowed the efficient detection of immobilized PBPs. Binding curves for BIO-AMP interaction with PBPs were then measured, and used to calculate apparent Km’s for each PBP’s interaction with BIO-AMP. Finally this assay was demonstrated for use in competition assays for the determination and characterization (KI) of unlabeled β–lactam PBP inhibitors.
MATERIALS and METHODS1
General
Biotin-ampicillin conjugate (BIO-AMP) was prepared by a modification of the method of Dargis and Malouin [18]. The PBPs used in this study were generous gifts from Professor Robert Nicholas (University of North Carolina).
PBP loading, labeling and detection
An ELISA-like protocol was used in black-walled Costar microtiter plates (Costar #3631), with gentle rocking at room temperature used for all steps. For PBP attachment, wells were treated with 2 μg of PBP in 50 μL of PBS/20% glycerol at 25 °C for 30 min, followed by treatment (3x) with 150 μL/well of blocking buffer (PBS/0.2% Tween-20), and then washed (3x) with 200 μL/well of washing buffer (PBS/0.05% Tween-20). To label PBPs in initial proof-of-principle experiments, 50 μL of 100 μM BIO-AMP in PBS was added to the wells. After 10 min the PBPs were denatured. Denaturation is necessary because PBPs catalyze the slow turnover of their β–lactam adducts, and loss of PBP-bound BIO-AMP would result in loss of signal. A number of denaturing conditions were tested, with heating at 80 °C for 3 min followed by quick cooling on ice giving the best results (data not shown). The plates were then washed (3x) with washing buffer. Streptavidin-horse radish peroxidase (HRP) conjugate (Pierce #21126) (50 μL of 0.1 μg/ml) was then added to each well. After 30 min the wells were washed (3x), and 100 μL of a fluorescent HRP substrate mixture (1 mM H2O2, 20 μM Amplex Red (Molecular Probes) in 100 mM Tris pH 8.5) was added to each well. After 60 min the fluorescence signal was read (Excitation: 546 nm, Emission: 595 nm).
Determination of BIO-AMP Km for binding vs various PBPs
PBPs turnover β–lactams (albeit usually very slowly). To assess the Km for BIO-AMP binding to a given PBP, the microtiter plate bound PBP was treated with serially diluted (steps of 5) concentrations of BIO-AMP, and the remaining steps of the assay performed as described above. Signals were plotted, and the set of 5 data points bracketing the midpoint of the saturation curve were analyzed for the Km of binding by fitting the data to Eqn. S7 (Supplementary Material).
| Eqn. S7 |
Application to HMM PBP-inhibitor screening and characterization
For inhibitor screening and characterization, BIO-AMP was used at a fixed concentration equal to the determined Km for a PBP. This is high enough to give 1/2 of the maximum possible signal and low enough to still allow inhibition to be readily detected. To demonstrate this capability, NG PBP2 was characterized for inhibition by ampicillin. NG PBP2 was first attached to the wells of a microtiter plate as described above. Competitive ampicillin/BIO-AMP binding was performed by adding 100 μL samples of serially (steps of 5) diluted solutions of ampicillin in the presence of 1.1 μM BIO-AMP (the Km for BIO-AMP vs NG PBP2, Table 1). After 15 minutes the binding reactions were stopped and developed as described above. With [BIO-AMP] = Km, and taking into account the background (blank) fluorescence, the inhibitor binding isotherm will be described by
| Eqn. S10 |
Inhibitor binding data were plotted, and the set of 5 data points bracketing the midpoint of the saturation curve were analyzed for the KI of binding by fitting the data to Eqn. S10.
Table 1.
Microtiter plate determined Km values for BIO-AMP with several PBPs.
| Enzyme |
Km (SE) (μM) |
|---|---|
| EC PBP1B | 1.6 (0.2) |
| NG PBP1 | 0.9 (0.2) |
| NG PBP2 | 1.1 (0.4) |
| NG PBP3 | 0.011 (0.003) |
| EC PBP5 | 100 (30) |
RESULTS and DISCUSSION
The key steps to this assay are PBP binding to the microtiter plate, and the BIO-AMP – PBP labeling reaction. The PBPs are generally not stable at room temperature for long periods, and for this reason incubations for loading PBPs onto the wells of microtiter plates were limited to 30 minutes. Also, since PBP–β-lactam complexes are turned over, it is necessary to “freeze” such a complex in a stable from by denaturing the PBP. At the same time, the PBP-β–lactam ester linkage is fairly labile, and the denaturation conditions could not too harsh. A number of alternative methods were tested for this step, with heating at 80 °C for three minutes giving the best results.
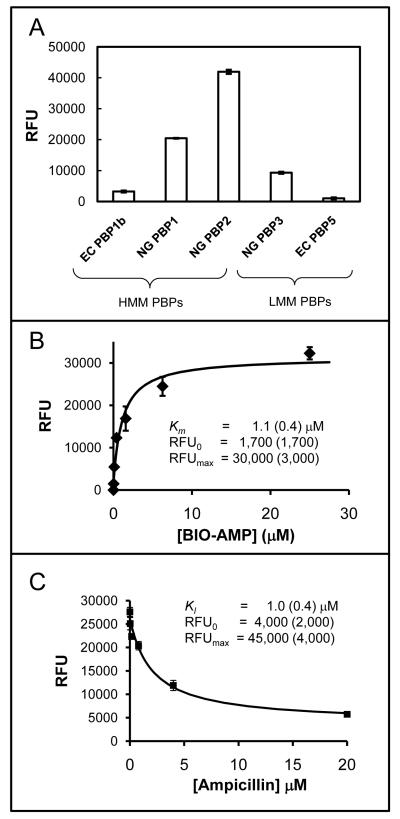
A preliminary test demonstrated readily detectible signals for all of the PBPs tested – both HMM and LMM (Fig. 1A). Substantial variation in the signal between PBPs was observed, presumably due to differences between PBPs in the efficiency of binding to the microtiter plates. Next, the dependence on BIO-AMP concentration was determined. An example of the fluorescence signal from NG PBP2 vs [BIO-AMP] is shown in Figure 1B, and a summary of Km values for all of the PBPs included in this study are given in Table 1. This demonstrated a wide range of values for the Km for individual PBPs, reflecting their differing affinities for the BIO-AMP reagent. Finally, to demonstrate the potential of this approach for the characterization of active site directed PBPs inhibitors, the competitive inhibition of NG PBP2 by ampicillin was characterized, as illustrated in Figure 1C. The KI obtained for ampicillin of 1.0 μM is very close to the Km obtained for BIO-AMP of 1.0 μM (Km and KI values are equivalent for slowly turned over substrates, as in the present case). These values are both close to the previously reported value of 0.5 μM for NG PBP2 binding to penicillin G determined using a classical approach based on radiolabeled penicillin G [21]. These results indicated that NG PBP2 is relatively insensitive to the differences between these three (ampicillin, BIO-AMP, and penicillin G) β-lactams.
Figure 1.
Panel A – Bar graph of the fluorescence reading (RFU) obtained from 5 different PBPs after loading, treatment with 100 mM BIO-AMP, and development as described in the text (+/- SE, n=4). Panel B – Plot of the RFU for NG PBP2 vs [BIO-AMP] (+/- SE, n=4). The best fit curve (Eqn. S7) and parameter values (+/- SE) are also shown. Panel C – RFU for NG PBP2 vs [AMP] at fixed [BIO-AMP] (equal to its Km) (+/- SE, n=4). The best fit curve (Eqn. S10) and parameter values are also shown.
The assay developed in this study provides a general method for screening and characterizing active site directed inhibitors for the lethal target HMM PBPs. Such an assay is expected to facilitate the development of new antibacterial agents targeting the HMM PBPs, and will also augment assays based on thiolester [7, 8], and lipid II [9-12] based substrates for studies on the biochemistry of the HMM PBPs.
Supplementary Material
Acknowledgements
Supported by the National Institutes of Health (Grant number GM60149 (WGG)), the American Heart Association (Grant number 0650179Z (WGG)), and by funds from the University of Missouri (University Research Board Grant K2303015 (WGG)).
Footnotes
Supplementary data (Detailed Materials and Methods) available on IDEAL (http://www.idealibrary.com).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- AMP
- ampicillin
- BIO
- biotin
- EC
- E. coli
- LMM
- low molecular mass
- HMM
- high molecular mass
- NG
- Neisseria gonorrhoeae
- PBP
- penicillin-binding protein
- PBS
- phosphate buffered saline
- RFU
- relative fluorescence units
References
- [1].Ghuysen JM. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- [2].Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–93. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev. 2006;30:673–91. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- [4].Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–58. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- [5].van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11:25R–36R. doi: 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]
- [6].Pratt RF. Substrate specificity of bacterial DD-peptidases (penicillin-binding proteins) Cell Mol Life Sci. 2008;65:2138–55. doi: 10.1007/s00018-008-7591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Adam M, Damblon C, Plaitin B, Christiaens L, Frere JM. Chromogenic depsipeptide substrates for beta-lactamases and penicillin-sensitive DD-peptidases. Biochem J. 1990;270:525–9. doi: 10.1042/bj2700525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Adam M, Damblon C, Jamin M, Zorzi W, Dusart V, Galleni M, el Kharroubi A, Piras G, Spratt BG, Keck W, et al. Acyltransferase activities of the high-molecular-mass essential penicillin-binding proteins. Biochem J. 1991;279(Pt 2):601–4. doi: 10.1042/bj2790601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Suzuki H, van Heijenoort Y, Tamura T, Mizoguchi J, Hirota Y, van Heijenoort J. In vitro peptidoglycan polymerization catalysed by penicillin binding protein 1b of Escherichia coli K-12. FEBS Lett. 1980;110:245–9. doi: 10.1016/0014-5793(80)80083-4. [DOI] [PubMed] [Google Scholar]
- [10].Nakagawa J, Matsuhashi S. Molecular divergence of a major peptidoglycan synthase with transglycosylase-transpeptidase activities in Eschericia coli. Biochem Biophys Res Commun. 1982;105 doi: 10.1016/0006-291x(82)90964-0. [DOI] [PubMed] [Google Scholar]
- [11].Ye XY, Lo MC, Brunner L, Walker D, Kahne D, Walker S. Better substrates for bacterial transglycosylases. Journal of the American Chemical Society. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- [12].Schwartz B, Markwalder JA, Wang Y. Lipid II: total synthesis of the bacterial cell wall precursor and utilization as a substrate for glycosyltransfer and transpeptidation by penicillin binding protein (PBP) 1b of Escherichia coli. J Am Chem Soc. 2001;123:11638–43. doi: 10.1021/ja0166848. [DOI] [PubMed] [Google Scholar]
- [13].van Heijenoort Y, Gomez M, Derrien M, Ayala J, van Heijenoort J. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J Bacteriol. 1992;174:3549–57. doi: 10.1128/jb.174.11.3549-3557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chandrakala B, Elias BC, Mehra U, Umapathy NS, Dwarakanath P, Balganesh TS, deSousa SM. Novel scintillation proximity assay for measuring membrane-associated steps of peptidoglycan biosynthesis in Escherichia coli. Antimicrob Agents Chemother. 2001;45:768–75. doi: 10.1128/AAC.45.3.768-775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chandrakala B, Shandil RK, Mehra U, Ravishankar S, Kaur P, Usha V, Joe B, deSousa SM. High-throughput screen for inhibitors of transglycosylase and/or transpeptidase activities of Escherichia coli penicillin binding protein 1b. Antimicrob Agents Chemother. 2004;48:30–40. doi: 10.1128/AAC.48.1.30-40.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jha RK, de Sousa SM. Microplate assay for inhibitors of the transpeptidase activity of PBP1b of Escherichia coli. J Biomol Screen. 2006;11:1005–14. doi: 10.1177/1087057106294364. [DOI] [PubMed] [Google Scholar]
- [17].Ramachandran V, Chandrakala B, Kumar VP, Usha V, Solapure SM, de Sousa SM. Screen for inhibitors of the coupled transglycosylase-transpeptidase of peptidoglycan biosynthesis in Escherichia coli. Antimicrob Agents Chemother. 2006;50:1425–32. doi: 10.1128/AAC.50.4.1425-1432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dargis M, Malouin F. Use of biotinylated beta-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria. Antimicrob Agents Chemother. 1994;38:973–80. doi: 10.1128/aac.38.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].DeLoney CR, Schiller NL. Competition of various beta-lactam antibiotics for the major penicillin-binding proteins of Helicobacter pylori: antibacterial activity and effects on bacterial morphology. Antimicrob Agents Chemother. 1999;43:2702–9. doi: 10.1128/aac.43.11.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bamberger DM, Herndon BL, Fitch J, Florkowski A, Parkhurst V. Effects of neutrophils on cefazolin activity and penicillin-binding proteins in Staphylococcus aureus abscesses. Antimicrob Agents Chemother. 2002;46:2878–84. doi: 10.1128/AAC.46.9.2878-2884.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schultz DE, Spratt BG, Nicholas RA. Expression and purification of a soluble form of penicillin-binding protein 2 from both penicillin-susceptible and penicillin-resistant Neisseria gonorrhoeae. Protein Expr Purif. 1991;2:339–49. doi: 10.1016/1046-5928(91)90092-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.