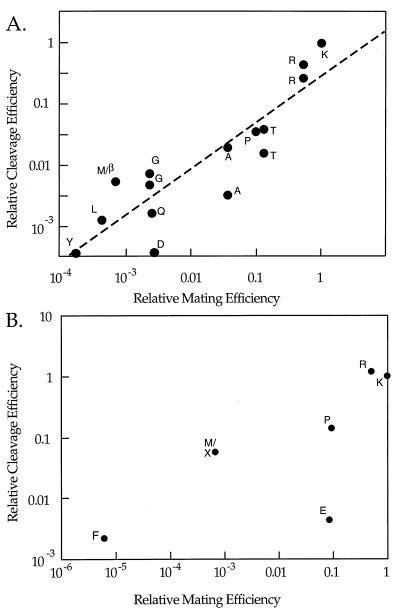
Figure 4.
In vitro vs. in vivo cleavage efficiency. (A) Peptidyl-MCA substrates. Relative efficiency of the peptidyl-MCA substrates is correlated with the in vivo-mating efficiency. Relative cleavage efficiency is defined as the ratio of kcat/KM for a peptidyl-MCA substrate with the indicated residue at P2 to the kcat/KM for a peptidyl-MCA substrate with Lys at P2. Data for kcat/KM of purified secreted, soluble Kex2 protease with peptidyl-MCA substrates are from Brenner and Fuller (7), Rockwell, et al. (9), Rockwell and Fuller (10), and N. Rockwell and R.S.F. (unpublished results). P2 residues of substrates compared are indicated in the single letter code (β represents nor-leucine). A substrate with nor-leucine at P2 was compared with mating efficiency with Met at P2 in vivo (YCpG-MFα1–110). In certain cases (Arg, Thr, Ala, and Gly), two synthetic substrates were available for comparison. The dashed line represents a line fit for direct proportionality between log (kcat/KM) and the log of the mating efficiency (correlation coefficient, r = 0.886; slope = 0.75). (B) Internally quenched fluorogenic peptide substrates. A correlation plot of relative efficiency of cleavage of the internally quenched fluorogenic peptide substrates (kcat/KM) v. relative mating efficiency for corresponding P2 substitutions. Relative cleavage efficiency is defined as the ratio of kcat/KM for an internally quenched fluorogenic peptide substrate with the indicated residue at P2 to the kcat/KM for an internally quenched fluorogenic peptide substrate with Lys at P2. Data for kcat/KM of purified secreted, soluble Kex2 protease with internally quenched fluorogenic peptide substrates are from ref. 9. P2 residues of substrates compared are indicated in the single letter code. The internally quenched fluorogenic peptides were of the sequence Arg-GlE-norLeu-Tyr-Xaa-Arg-↓-Glu-Ala-Glu-Ala-LyD-Arg, in which “GlE” represents glutamyl-EDANS, [EDANS, 5-(2-aminoethyl-amino)-naphthalene-1-sulfonic acid], “Xaa” represents the particular P2 substitution, and “LyD” represents lysyl-DABSYL (DABCYL, 4-(4-dimethylaminophenyl)-azobenzoyl) (41, 42). This sequence was based on the Kex2 cleavage site between the α-factor repeats in pro-α-factor. A substrate with nor-leucine (X) at P2 was compared with mating efficiency with Met at P2 in vivo (YCpG-MFα1–110).