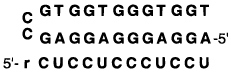Table 4.
The Effect of Ligand Structure on the Binding of an RNA Strand (Sequence rCUCCUCCCUCCU) Shown Are Melting Transition Temperatures (Tm (°C)) and Free Energies (−ΔG° (kcal/mol)) at pH 7.0a
Conditions: 100 mM NaCl, 10 mM MgCl2, 10 mM Na•PIPES, pH 7.0, 1.5 µM each DNA strand.
Error in Tm values and in free energies are estimated at ±1.0 °C and ±10%, respectively.